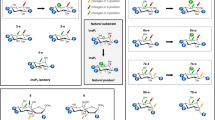
Abstract
Inositol pyrophosphates (such as IP7 and IP8) are multifunctional signaling molecules that regulate diverse cellular activities. Inositol pyrophosphates have 'high-energy' phosphoanhydride bonds, so their enzymatic synthesis requires that a substantial energy barrier to the transition state be overcome. Additionally, inositol pyrophosphate kinases can show stringent ligand specificity, despite the need to accommodate the steric bulk and intense electronegativity of nature's most concentrated three-dimensional array of phosphate groups. Here we examine how these catalytic challenges are met by describing the structure and reaction cycle of an inositol pyrophosphate kinase at the atomic level. We obtained crystal structures of the kinase domain of human PPIP5K2 complexed with nucleotide cofactors and either substrates, product or a MgF3− transition-state mimic. We describe the enzyme's conformational dynamics, its unprecedented topological presentation of nucleotide and inositol phosphate, and the charge balance that facilitates partly associative in-line phosphoryl transfer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Streb, H., Irvine, R.F., Berridge, M.J. & Schulz, I. Release of Ca2+ from a nonmitochondrial store in pancreatic cells by inositol-1,4,5-trisphosphate. Nature 306, 67–69 (1983).
Monserrate, J.P. & York, J.D. Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22, 365–373 (2010).
Stephens, L. et al. The detection, purification, structural characterization and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s). J. Biol. Chem. 268, 4009–4015 (1993).
Menniti, F.S., Miller, R.N., Putney, J.W. Jr. & Shears, S.B. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 268, 3850–3856 (1993).
Barker, C.J., Illies, C., Gaboardi, G.C. & Berggren, P.O. Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 66, 3851–3871 (2009).
Burton, A., Hu, X. & Saiardi, A. Are inositol pyrophosphates signalling molecules? J. Cell. Physiol. 220, 8–15 (2009).
Shears, S.B. Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 76, 236–252 (2009).
Prasad, A. et al. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 12, 752–760 (2011).
Chakraborty, A. et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910 (2010).
Lee, Y.S., Huang, K., Quiocho, F.A. & O'Shea, E.K. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4, 25–32 (2008).
Saiardi, A. et al. Inositol pyrophosphate: physiologic phosphorylation of proteins. Science 306, 2101–2105 (2004).
Hand, C.E. & Honek, J.F. Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: a semi-empirical investigation. Bioorg. Med. Chem. Lett. 17, 183–188 (2007).
Choi, J.H., Williams, J., Cho, J., Falck, J.R. & Shears, S.B. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 282, 30763–30775 (2007).
Gokhale, N.A., Zaremba, A. & Shears, S.B. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem. J. 434, 415–426 (2011).
Saiardi, A., Nagata, E., Luo, H.R., Snowman, A.M. & Snyder, S.H. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 276, 39179–39185 (2001).
Fridy, P.C., Otto, J.C., Dollins, D.E. & York, J.D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 282, 30754–30762 (2007).
Saiardi, A., Erdjument-Bromage, H., Snowman, A., Tempst, P. & Snyder, S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326 (1999).
Mulugu, S. et al. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 (2007).
Lin, H. et al. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J. Biol. Chem. 284, 1863–1872 (2009).
Draškovič, P. et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 15, 274–286 (2008).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Lee, D., Redfern, O. & Orengo, C. Predicting protein function from sequence and structure. Nat. Rev. Mol. Cell Biol. 8, 995–1005 (2007).
Miller, G.J., Wilson, M.P., Majerus, P.W. & Hurley, J.H. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol. Cell 18, 201–212 (2005).
Chamberlain, P.P. et al. Integration of inositol phosphate signaling pathways via human ITPK1. J. Biol. Chem. 282, 28117–28125 (2007).
Shears, S.B. How versatile are inositol phosphate kinases? Biochem. J. 377, 265–280 (2004).
Holmes, W. & Jogl, G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J. Biol. Chem. 281, 38109–38116 (2006).
Herschlag, D. The role of induced fit and conformational changes in specificity and catalysis. Bioorg. Chem. 16, 62–96 (1988).
Graham, D.L. et al. MgF3− as a transition state analog of phosphoryl transfer. Chem. Biol. 9, 375–381 (2002).
Mildvan, A.S. Mechanisms of signaling and related enzymes. Proteins 29, 401–416 (1997).
Cannon, W.R., Singleton, S.F. & Benkovic, S.J. A perspective on biological catalysis. Nat. Struct. Biol. 3, 821–833 (1996).
Miller, G.J. & Hurley, J.H. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol. Cell 15, 703–711 (2004).
Gonzalez, B. et al. Structure of a human inositol 1,4,5-trisphosphate 3-kinase; substrate binding reveals why it is not a phosphoinositide 3-kinase. Mol. Cell 15, 689–701 (2004).
González, B., Banos-Sanz, J.I., Villate, M., Brearley, C.A. & Sanz-Aparicio, J. Inositol 1,3,4,5,6-pentakisphosphate 2-kinase is a distant IPK member with a singular inositide binding site for axial 2-OH recognition. Proc. Natl. Acad. Sci. USA 107, 9608–9613 (2010).
Madhusudan, Xuong, N.H. & Taylor, S.S. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat. Struct. Biol. 9, 273–277 (2002).
Reddy, K.M., Reddy, K.K. & Falck, J.R. Synthesis of 2- and 5-diphospho-myo-inositol pentakisphosphate (2- and 5-PP-InsP5). Tetrahedron Lett. 38, 4951–4952 (1997).
Albert, C. et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem. J. 327, 553–560 (1997).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Winn, M.D., Murshudov, G.N. & Papiz, M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003).
Acknowledgements
Data were collected at the Southeast Regional Collaborative Access Team 22-ID/22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. The use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. This work was supported by the Intramural Research Program of the National Institutes of Health and National Institute of Environmental Health Sciences (NIEHS). We are grateful to L.C. Pedersen for advice and support. We also thank H. Ke for his assistance in writing the manuscript. Expression vectors were prepared by the NIEHS Protein Expression Core Facility.
Author information
Authors and Affiliations
Contributions
H.W., T.M.T.H. and S.B.S. designed experiments and wrote the manuscript. H.W. and S.B.S. performed the experiments. J.R.F. synthesized substrate for hPPIP5K2.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 3747 kb)
Supplementary Movie 1
Wang_SuppMovie1.mov (MOV 5167 kb)
Supplementary Movie 2
Wang_SuppMovie2.mov (MOV 3991 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Falck, J., Hall, T. et al. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol 8, 111–116 (2012). https://doi.org/10.1038/nchembio.733
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.733
This article is cited by
-
Substrate promiscuity of inositol 1,4,5-trisphosphate kinase driven by structurally-modified ligands and active site plasticity
Nature Communications (2024)
-
Functions, Mechanisms, and therapeutic applications of the inositol pyrophosphates 5PP-InsP5 and InsP8 in mammalian cells
Journal of Cardiovascular Translational Research (2023)
-
Exploring the interaction mechanism between antagonist and the jasmonate receptor complex by molecular dynamics simulation
Journal of Computer-Aided Molecular Design (2022)
-
Plant phosphate nutrition: sensing the stress
Stress Biology (2022)
-
PPIP5K2 promotes colorectal carcinoma pathogenesis through facilitating DNA homologous recombination repair
Oncogene (2021)