Abstract
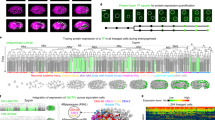
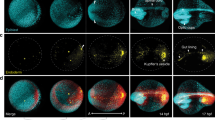
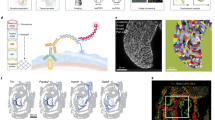
Transcription factors have diverse roles during embryonic development, combinatorially controlling cellular states in a spatially and temporally defined manner. Resolving the dynamic transcriptional profiles that underlie these patterning processes is essential for understanding embryogenesis at the molecular level. Here we show how temporal, tissue-specific changes in embryonic transcription factor function can be discerned by integrating caged morpholino oligonucleotides with photoactivatable fluorophores, fluorescence-activated cell sorting and microarray technologies. As a proof of principle, we have dynamically profiled No tail a (Ntla)-dependent genes at different stages of axial mesoderm development in zebrafish, discovering discrete sets of transcripts that are coincident with either notochord cell fate commitment or differentiation. Our studies reveal new regulators of notochord development and the sequential activation of distinct transcriptomes within a cell lineage by a single transcriptional factor and demonstrate how optically controlled chemical tools can dissect developmental processes with spatiotemporal precision.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ouyang, X. & Chen, J.K. Synthetic strategies for studying embryonic development. Chem. Biol. 17, 590–606 (2010).
Shestopalov, I.A., Sinha, S. & Chen, J.K. Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol. 3, 650–651 (2007).
Ouyang, X. et al. Versatile synthesis and rational design of caged morpholinos. J. Am. Chem. Soc. 131, 13255–13269 (2009).
Tang, X., Maegawa, S., Weinberg, E.S. & Dmochowski, I.J. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc. 129, 11000–11001 (2007).
Tomasini, A.J., Schuler, A.D., Zebala, J.A. & Mayer, A.N. PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis 47, 736–743 (2009).
Deiters, A. et al. Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc. 132, 15644–15650 (2010).
Schulte-Merker, S., Ho, R.K., Herrmann, B.G. & Nusslein-Volhard, C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116, 1021–1032 (1992).
Halpern, M.E., Ho, R.K., Walker, C. & Kimmel, C.B. Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell 75, 99–111 (1993).
Schulte-Merker, S., van Eeden, F.J., Halpern, M.E., Kimmel, C.B. & Nusslein-Volhard, C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 120, 1009–1015 (1994).
Halpern, M.E. et al. Genetic interactions in zebrafish midline development. Dev. Biol. 187, 154–170 (1997).
Amacher, S.L., Draper, B.W., Summers, B.R. & Kimmel, C.B. The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development 129, 3311–3323 (2002).
Melby, A.E., Warga, R.M. & Kimmel, C.B. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development 122, 2225–2237 (1996).
Garnett, A.T. et al. Identification of direct T-box target genes in the developing zebrafish mesoderm. Development 136, 749–760 (2009).
Goering, L.M. et al. An interacting network of T-box genes directs gene expression and fate in the zebrafish mesoderm. Proc. Natl. Acad. Sci. USA 100, 9410–9415 (2003).
Morley, R.H. et al. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. USA 106, 3829–3834 (2009).
Talbot, W.S. et al. A homeobox gene essential for zebrafish notochord development. Nature 378, 150–157 (1995).
Ochi, H., Hans, S. & Westerfield, M. Smarcd3 regulates the timing of zebrafish myogenesis onset. J. Biol. Chem. 283, 3529–3536 (2008).
Showell, C., Binder, O. & Conlon, F.L. T-box genes in early embryogenesis. Dev. Dyn. 229, 201–218 (2004).
Moulton, J.D. & Yan, Y.L. Using Morpholinos to control gene expression. Curr. Protoc. Mol. Biol. 83, 26.8.1–26.8.29 (2008).
Gajewski, M. et al. Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130, 4269–4278 (2003).
Conlon, F.L., Sedgwick, S.G., Weston, K.M. & Smith, J.C. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development 122, 2427–2435 (1996).
Gansner, J.M. & Gitlin, J.D. Essential role for the alpha 1 chain of type VIII collagen in zebrafish notochord formation. Dev. Dyn. 237, 3715–3726 (2008).
Stemple, D.L. et al. Mutations affecting development of the notochord in zebrafish. Development 123, 117–128 (1996).
Currie, P.D. & Ingham, P.W. Induction of a specific muscle cell type by a hedgehog-like protein in zebrafish. Nature 382, 452–455 (1996).
Chen, J.K., Taipale, J., Cooper, M.K. & Beachy, P.A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002).
Hill, M.M. et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 (2008).
Nixon, S.J. et al. Caveolin-1 is required for lateral line neuromast and notochord development. J. Cell Sci. 120, 2151–2161 (2007).
Nixon, S.J. et al. Zebrafish as a model for caveolin-associated muscle disease; caveolin-3 is required for myofibril organization and muscle cell patterning. Hum. Mol. Genet. 14, 1727–1743 (2005).
Franchi-Gazzola, R. et al. The role of the neutral amino acid transporter SNAT2 in cell volume regulation. Acta Physiol. (Oxf.) 187, 273–283 (2006).
Glickman, N.S., Kimmel, C.B., Jones, M.A. & Adams, R.J. Shaping the zebrafish notochord. Development 130, 873–887 (2003).
Draper, B.W., Stock, D.W. & Kimmel, C.B. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development 130, 4639–4654 (2003).
Chesley, P. Development of the short-tailed mutant in the house mouse. J. Exp. Zool. 70, 429–459 (1935).
Wilkinson, D.G., Bhatt, S. & Herrmann, B.G. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343, 657–659 (1990).
Melby, A.E., Kimelman, D. & Kimmel, C.B. Spatial regulation of floating head expression in the developing notochord. Dev. Dyn. 209, 156–165 (1997).
Brand, M., Granato, M. & Nusslein-Volhard, C. Keeping and raising zebrafish. in Zebrafish: A Practical Approach (eds. Nusslein-Volhard, C. & Dahm, R.) 7–37 (Oxford University Press, 2002).
Bill, B.R., Petzold, A.M., Clark, K.J., Schimmenti, L.A. & Ekker, S.C. A primer for morpholino use in zebrafish. Zebrafish 6, 69–77 (2009).
Jowett, T. Double in situ hybridization techniques in zebrafish. Methods 23, 345–358 (2001).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Acknowledgements
We thank S. Amacher and A. Garnett for helpful discussions, M. Halpern (Carnegie Institution) for providing ntla and flh cDNA, W. Talbot (Stanford University) for pax2a cDNA, J. Mich and X. Ouyang (both from Stanford University) for sharing in situ hybridization probes and C. Crumpton for technical assistance with FACS. This work was supported by funding from the US National Institutes of Health (R01 GM072600, R01 GM087292 and DP1 OD003792) and the March of Dimes Foundation (1-FY-08-433).
Author information
Authors and Affiliations
Contributions
J.K.C. and I.A.S. conceived the study; J.K.C. directed its execution; I.A.S. and C.L.W.P. designed, conducted and interpreted the experiments; and J.K.C. and I.A.S. wrote the manuscript with contributions from C.L.W.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 4178 kb)
Rights and permissions
About this article
Cite this article
Shestopalov, I., Pitt, C. & Chen, J. Spatiotemporal resolution of the Ntla transcriptome in axial mesoderm development. Nat Chem Biol 8, 270–276 (2012). https://doi.org/10.1038/nchembio.772
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.772
This article is cited by
-
Illuminating developmental biology through photochemistry
Nature Chemical Biology (2017)
-
Tbx16 regulates hox gene activation in mesodermal progenitor cells
Nature Chemical Biology (2016)