Abstract
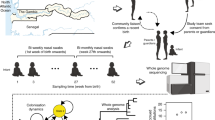
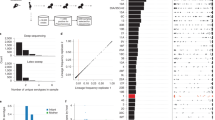
Streptococcus pneumoniae ('pneumococcus') causes an estimated 14.5 million cases of serious disease and 826,000 deaths annually in children under 5 years of age1. The highly effective introduction of the PCV7 pneumococcal vaccine in 2000 in the United States2,3 provided an unprecedented opportunity to investigate the response of an important pathogen to widespread, vaccine-induced selective pressure. Here, we use array-based sequencing of 62 isolates from a US national monitoring program to study five independent instances of vaccine escape recombination4, showing the simultaneous transfer of multiple and often large (up to at least 44 kb) DNA fragments. We show that one such new strain quickly became established, spreading from east to west across the United States. These observations clarify the roles of recombination and selection in the population genomics of pneumococcus and provide proof of principle of the considerable value of combining genomic and epidemiological information in the surveillance and enhanced understanding of infectious diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
O'Brien, K.L. et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902 (2009).
Whitney, C.G. et al. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348, 1737–1746 (2003).
Pilishvili, T. et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201, 32–41 (2010).
Brueggemann, A.B., Pai, R., Crook, D.W. & Beall, B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3, e168 (2007).
Eskola, J. et al. A randomized, prospective field trial of a conjugate vaccine in the protection of infants and young children against invasive Haemophilus influenzae type b disease. N. Engl. J. Med. 323, 1381–1387 (1990).
Perkins, B.A. New opportunities for prevention of meningococcal disease. J. Am. Med. Assoc. 283, 2842–2843 (2000).
Campbell, H., Borrow, R., Salisbury, D. & Miller, E. Meningococcal C conjugate vaccine: the experience in England and Wales. Vaccine 27 (suppl. 2), B20–B29 (2009).
Lipsitch, M. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 5, 336–345 (1999).
Spratt, B.G. & Greenwood, B.M. Prevention of pneumococcal disease by vaccination: does serotype replacement matter? Lancet 356, 1210–1211 (2000).
Bogaert, D. et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 363, 1871–1872 (2004).
Beall, B. et al. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44, 999–1017 (2006).
Moore, M.R. et al. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197, 1016–1027 (2008).
Enright, M.C. & Spratt, B.G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144, 3049–3060 (1998).
Pai, R. et al. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192, 1988–1995 (2005).
Griffith, F. The significance of pneumococcal types. J. Hyg. (Lond.) 27, 113–159 (1928).
Avery, O.T., Macleod, C.M. & McCarty, M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus Type III. J. Exp. Med. 79, 137–158 (1944).
Lacks, S. & Hotchkiss, R.D. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39, 508–518 (1960).
Trzciński, K., Thompson, C.M. & Lipsitch, M. Single-step capsular transformation and acquisition of penicillin resistance in Streptococcus pneumoniae. J. Bacteriol. 186, 3447–3452 (2004).
Hiller, N.L. et al. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6, e1001108 (2010).
Croucher, N.J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011).
Feil, E.J., Smith, J.M., Enright, M.C. & Spratt, B.G. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154, 1439–1450 (2000).
Tettelin, H. et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 (2001).
Cutler, D.J. et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 11, 1913–1925 (2001).
Zwick, M.E. et al. Microarray-based resequencing of multiple Bacillus anthracis isolates. Genome Biol. 6, R10 (2005).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Darling, A.C., Mau, B., Blattner, F.R. & Perna, N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2007).
Acknowledgements
The authors gratefully acknowledge the clinicians, microbiologists and investigators of the ABCs program of the Emerging Infections Program Network. We thank X. Didelot for contributions to the analysis of Illumina genomic data. This work was funded by the Wellcome Trust (079126/Z/06/Z). T.E.A.P. and D.W.C. are funded by the UK National Health Service (NHS) National Institute for Health Research (NIHR) Oxford Biomedical Research Centre and NIHR Senior Investigator Awards. P.D. is funded by a Wellcome Trust Core Award Grant (090532/Z/09/Z) and is supported, in part, by a Wolfson Royal Society Merit Award. R.B. is supported by the NHS NIHR Oxford Biomedical Research Centre and the UK Clinical Research Collaboration (UKCRC) (MRC UK G0800778 and Wellcome Trust 087646/2/08/Z). A.B.B. is a Wellcome Trust Career Development Fellow (083511/Z/07/Z). Genetic and epidemiological data are available from the authors.
Author information
Authors and Affiliations
Contributions
B.B. and R.E.G. collected isolates. A.B.B., B.B., R.B., D.W.C., P.D., T.G., R.M.H. and T.E.A.P. planned experiments. A.B.B., B.B., R.L.-G., M.R.M. and T.E.A.P. collected and analyzed epidemiological data. A.B.B., R.E.G., T.G. and T.S. performed molecular typing. T.G., T.H. and T.S. prepared DNA and performed microarray-based sequencing. T.G., A.B.B., R.B., P.D., C.C.A.S. and E.G. analyzed molecular data. R.B., P.D. and D.W.C. wrote the manuscript. A.B.B., B.B., T.G., M.R.M. and T.E.A.P. contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.C. and T.P. are in receipt of a research grant for pneumococcal surveillance from Pfizer. A.B.B. is in receipt of grant funding from GlaxoSmithKline Biologicals and Pfizer (Wyeth) Vaccines.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–8, Supplementary Figures 1–3 and Supplementary Note (PDF 588 kb)
Rights and permissions
About this article
Cite this article
Golubchik, T., Brueggemann, A., Street, T. et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet 44, 352–355 (2012). https://doi.org/10.1038/ng.1072
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1072
This article is cited by
-
Comparative genomic analysis of two ST320 Streptococcus pneumoniae isolates, representing serotypes 19A and 19F
BMC Genomic Data (2023)
-
A novel invasive Streptococcus pyogenes variant sublineage derived through recombinational replacement of the emm12 genomic region
Scientific Reports (2023)
-
Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape
Nature (2022)
-
Identification of evolutionarily conserved virulence factor by selective pressure analysis of Streptococcus pneumoniae
Communications Biology (2019)
-
Genomic analyses of pneumococci reveal a wide diversity of bacteriocins – including pneumocyclicin, a novel circular bacteriocin
BMC Genomics (2015)