Abstract
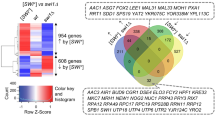
SWI/SNF, an evolutionarily conserved ATP-dependent chromatin-remodeling complex, has an important role in transcriptional regulation1. In Saccharomyces cerevisiae, SWI/SNF regulates the expression of ∼6% of total genes through activation or repression2. Swi1, a subunit of SWI/SNF, contains an N-terminal region rich in glutamine and asparagine, a notable feature shared by all characterized yeast prions—a group of unique proteins capable of self-perpetuating changes in conformation and function3. Here we provide evidence that Swi1 can become a prion, [SWI+]. Swi1 aggregates in [SWI+] cells but not in nonprion cells. Cells bearing [SWI+] show a partial loss-of-function phenotype of SWI/SNF. [SWI+] can be eliminated by guanidine hydrochloride treatment, HSP104 deletion or loss of Swi1. Moreover, we show [SWI+] is dominantly and cytoplasmically transmitted. Our findings reveal a novel mechanism of 'protein-only' inheritance that results in modification of chromatin-remodeling and, ultimately, global gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Martens, J.A. & Winston, F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13, 136–142 (2003).
Sudarsanam, P., Iyer, V.R., Brown, P.O. & Winston, F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97, 3364–3369 (2000).
Tuite, M.F. & Cox, B.S. Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 4, 878–890 (2003).
Wickner, R.B. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 (1994).
Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 95, 13363–13383 (1998).
Cox, B. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity 20, 505–521 (1965).
Lacroute, F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 206, 519–522 (1971).
Sondheimer, N. & Lindquist, S. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172 (2000).
Derkatch, I.L., Bradley, M.E., Hong, J.Y. & Liebman, S.W. Prions affect the appearance of other prions: the story of [PIN+]. Cell 106, 171–182 (2001).
Uptain, S.M. & Lindquist, S. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56, 703–741 (2002).
Chernoff, Y.O., Derkach, I.L. & Inge-Vechtomov, S.G. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 24, 268–270 (1993).
Masison, D.C. & Wickner, R.B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93–95 (1995).
Osherovich, L.Z. & Weissman, J.S. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 106, 183–194 (2001).
Derkatch, I.L., Bradley, M.E., Zhou, P., Chernoff, Y.O. & Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 (1997).
Derkatch, I.L. et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI.] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 101, 12934–12939 (2004).
Michelitsch, M.D. & Weissman, J.S. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 97, 11910–11915 (2000).
Saha, A., Wittmeyer, J. & Cairns, B.R. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7, 437–447 (2006).
Smith, C.L., Horowitz-Scherer, R., Flanagan, J.F., Woodcock, C.L. & Peterson, C.L. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10, 141–145 (2003).
Neigeborn, L. & Carlson, M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858 (1984).
Guidi, C.J. et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21, 3598–3603 (2001).
Roberts, C.W. & Orkin, S.H. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer 4, 133–142 (2004).
Ross, E.D., Minton, A. & Wickner, R.B. Prion domains: sequences, structures and interactions. Nat. Cell Biol. 7, 1039–1044 (2005).
Tuite, M.F., Mundy, C.R. & Cox, B.S. Agents that cause a high frequency of genetic change from [PSI+] to [psi−] in Saccharomyces cerevisiae. Genetics 98, 691–711 (1981).
Neigeborn, L. & Carlson, M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115, 247–253 (1987).
Li, L. & Lindquist, S. Creating a protein-based element of inheritance. Science 287, 661–664 (2000).
Parsell, D.A., Kowal, A.S., Singer, M.A. & Lindquist, S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478 (1994).
Conde, J. & Fink, G.R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA 73, 3651–3655 (1976).
Krobitsch, S., & Lindquist, S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA 97, 1589–1594 (2000).
Park, K.W., Hahn, J.S., Fan, Q., Thiele, D.J. & Li, L. De novo appearance and “strain” formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor. Genetics 173, 35–47 (2006).
Laurent, B.C., Treitel, M.A. & Carlson, M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol. Cell. Biol. 10, 5616–5625 (1990).
Acknowledgements
The authors thank M. Carlson (Department of Genetics and Development, Columbia University) for the gift of the pLS7 plasmid; B.C. Laurent (Department of Oncological Sciences, Mount Sinai School of Medicine) for the gift of the pLY14 plasmid; S. Lindquist (Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute of Technology and Howard Hughes Medical Institute) for the Hsp104 antibody; J. Workman for helpful discussions; E. Crow and G.E. Kim for technical assistance; R. Lawrence, C. Kowalczyk, R. Miller, T. Volpe, C. Long and E. Crow for critical comments and manuscript editing. This work was partially supported by grants from the United States Army (0850-370-R744), the Ellison Medical Foundation and the US National Institutes of Health (R01NS056086) to L.L.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Figures 1–6 (PDF 304 kb)
Rights and permissions
About this article
Cite this article
Du, Z., Park, KW., Yu, H. et al. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 40, 460–465 (2008). https://doi.org/10.1038/ng.112
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.112
This article is cited by
-
Prions in Microbes: The Least in the Most
Journal of Microbiology (2023)
-
Protein assembly systems in natural and synthetic biology
BMC Biology (2020)
-
Prions: Roles in Development and Adaptive Evolution
Journal of Molecular Evolution (2020)
-
Three J-proteins impact Hsp104-mediated variant-specific prion elimination: a new critical role for a low-complexity domain
Current Genetics (2020)
-
A viral expression factor behaves as a prion
Nature Communications (2019)