Abstract
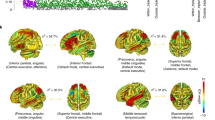
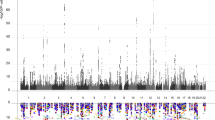
Identifying genetic variants influencing human brain structures may reveal new biological mechanisms underlying cognition and neuropsychiatric illness. The volume of the hippocampus is a biomarker of incipient Alzheimer's disease1,2 and is reduced in schizophrenia3, major depression4 and mesial temporal lobe epilepsy5. Whereas many brain imaging phenotypes are highly heritable6,7, identifying and replicating genetic influences has been difficult, as small effects and the high costs of magnetic resonance imaging (MRI) have led to underpowered studies. Here we report genome-wide association meta-analyses and replication for mean bilateral hippocampal, total brain and intracranial volumes from a large multinational consortium. The intergenic variant rs7294919 was associated with hippocampal volume (12q24.22; N = 21,151; P = 6.70 × 10−16) and the expression levels of the positional candidate gene TESC in brain tissue. Additionally, rs10784502, located within HMGA2, was associated with intracranial volume (12q14.3; N = 15,782; P = 1.12 × 10−12). We also identified a suggestive association with total brain volume at rs10494373 within DDR2 (1q23.3; N = 6,500; P = 5.81 × 10−7).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jack, C.R. Jr. et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer's disease. Alzheimers Dement. 7, 474–485 e4 (2011).
Simić, G., Kostovic, I., Winblad, B. & Bogdanovic, N. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. J. Comp. Neurol. 379, 482–494 (1997).
Wright, I.C. et al. Meta-analysis of regional brain volumes in schizophrenia. Am. J. Psychiatry 157, 16–25 (2000).
Videbech, P. & Ravnkilde, B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am. J. Psychiatry 161, 1957–1966 (2004).
Keller, S.S. & Roberts, N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia 49, 741–757 (2008).
Peper, J.S., Brouwer, R.M., Boomsma, D.I., Kahn, R.S. & Hulshoff Pol, H.E. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum. Brain Mapp. 28, 464–473 (2007).
Kremen, W.S. et al. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage 49, 1213–1223 (2010).
Maguire, E.A. et al. Navigation-related structural change in the hippocampi of taxi drivers. Proc. Natl. Acad. Sci. USA 97, 4398–4403 (2000).
Burgess, N., Maguire, E.A. & O'Keefe, J. The human hippocampus and spatial and episodic memory. Neuron 35, 625–641 (2002).
Snyder, J.S., Soumier, A., Brewer, M., Pickel, J. & Cameron, H.A. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature 476, 458–461 (2011).
Freitag, C.M. et al. Total brain volume and corpus callosum size in medication-naive adolescents and young adults with autism spectrum disorder. Biol. Psychiatry 66, 316–319 (2009).
Stanfield, A.C. et al. Towards a neuroanatomy of autism: a systematic review and meta-analysis of structural magnetic resonance imaging studies. Eur. Psychiatry 23, 289–299 (2008).
Posthuma, D. et al. The association between brain volume and intelligence is of genetic origin. Nat. Neurosci. 5, 83–84 (2002).
Fears, S.C. et al. Identifying heritable brain phenotypes in an extended pedigree of vervet monkeys. J. Neurosci. 29, 2867–2875 (2009).
Rogers, J. et al. On the genetic architecture of cortical folding and brain volume in primates. Neuroimage 53, 1103–1108 (2010).
Patenaude, B., Smith, S.M., Kennedy, D.N. & Jenkinson, M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56, 907–922 (2011).
Fischl, B. et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002).
Buckner, R.L. et al. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage 23, 724–738 (2004).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010).
Han, B. & Eskin, E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88, 586–598 (2011).
Bis, J.C. et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat. Genet. published online (15 April 2012; doi:10.1038/ng.2237).
Ripke, S. et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 43, 969–976 (2011).
Sklar, P. et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011).
Li, M.X., Gui, H.S., Kwan, J.S. & Sham, P.C. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 88, 283–293 (2011).
Bao, Y. et al. Expression and evolutionary conservation of the tescalcin gene during development. Gene expression patterns. Gene Exp. Patterns 9, 273–281 (2009).
Baumgartner, M., Patel, H. & Barber, D.L. Na+/H+ exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. Am. J. Physiol. Cell Physiol. 287, C844–C850 (2004).
Slepkov, E.R., Rainey, J.K., Sykes, B.D. & Fliegel, L. Structural and functional analysis of the Na+/H+ exchanger. Biochem. J. 401, 623–633 (2007).
Levay, K. & Slepak, V.Z. Tescalcin is an essential factor in megakaryocytic differentiation associated with Ets family gene expression. J. Clin. Invest. 117, 2672–2683 (2007).
Levay, K. & Slepak, V.Z. Up- or downregulation of tescalcin in HL-60 cells is associated with their differentiation to either granulocytic or macrophage-like lineage. Exp. Cell Res. 316, 1254–1262 (2010).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Gudbjartsson, D.F. et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 40, 609–615 (2008).
Sanna, S. et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 40, 198–203 (2008).
Weedon, M.N. et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 40, 575–583 (2008).
Fusco, A. & Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910 (2007).
Litterman, N. et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 9, e1001060 (2011).
Hammond, S.M. & Sharpless, N.E. HMGA2, microRNAs, and stem cell aging. Cell 135, 1013–1016 (2008).
Wright, M.J. & Martin, N.G. Brisbane adolescent twin study: outline of study methods and research projects. Aust. J. Psychol. 56, 65–78 (2004).
Jackson, D.N. MAB: Multidimensional Aptitude Battery Manual (Research Psychologists Press, Port Huron, Michigan, 1984).
Vogel, W. Discoidin domain receptors: structural relations and functional implications. FASEB J. 13 (suppl), S77–S82 (1999).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Johnson, M.B. et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009).
Smith, S.M. et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 (suppl 1), S208–S219 (2004).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001).
Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002).
Morey, R.A. et al. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage 45, 855–866 (2009).
Pardoe, H.R., Pell, G.S., Abbott, D.F. & Jackson, G.D. Hippocampal volume assessment in temporal lobe epilepsy: how good is automated segmentation? Epilepsia 50, 2586–2592 (2009).
Morey, R.A. et al. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum. Brain Mapp. 31, 1751–1762 (2010).
Pantel, J. et al. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus 10, 752–758 (2000).
Morra, J.H. et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. Neuroimage 43, 59–68 (2008).
Almasy, L. & Blangero, J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62, 1198–1211 (1998).
Purcell, S., Cherny, S.S. & Sham, P.C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19, 149–150 (2003).
Pei, Y.F., Li, J., Zhang, L., Papasian, C.J. & Deng, H.W. Analyses and comparison of accuracy of different genotype imputation methods. PLoS ONE 3, e3551 (2008).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Allen, A.S., Martin, E.R., Qin, X. & Li, Y.J. Genetic association tests based on ranks (GATOR) for quantitative traits with and without censoring. Genet. Epidemiol. 30, 248–258 (2006).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Browning, B.L. & Browning, S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Durston, S. et al. Differential effects of DRD4 and DAT1 genotype on fronto-striatal gray matter volumes in a sample of subjects with attention deficit hyperactivity disorder, their unaffected siblings, and controls. Mol. Psychiatry 10, 678–685 (2005).
Dick, D.M. et al. Genome-wide association study of conduct disorder symptomatology. Mol. Psychiatry 16, 800–808 (2011).
Stein, J.L. et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol. Psychiatry 16, 927–937, 881 (2011).
Chen, W.M. & Abecasis, G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 81, 913–926 (2007).
Aulchenko, Y.S., Struchalin, M.V. & van Duijn, C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Li, M.X., Sham, P.C., Cherny, S.S. & Song, Y.Q. A knowledge-based weighting framework to boost the power of genome-wide association studies. PLoS ONE 5, e14480 (2010).
Li, J. & Ji, L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005).
Kasperaviciūte, D. et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain 133, 2136–2147 (2010).
Trabzuni, D. et al. Quality control parameters on a large dataset of regionally-dissected human control brains for whole genome expression studies. J. Neurochem. 119, 275–282 (2011).
Heinzen, E.L. et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 6, e1 (2008).
Acknowledgements
Some authors received commercial funding unrelated to the topic of this paper. N.J.v.d.W. received speaking fees from Eli Lilly & Company and Wyeth and served on advisory panels of Eli Lilly & Company, Pfizer, Wyeth and Servier. A.A. received an investigator-initiated unrestricted research grant from Bristol-Myers Squibb and speaker's fees from AstraZeneca, Bristol-Myers Squibb and GlaxoSmithKline. H.J.G. received external research support from the German Research Foundation, the Federal Ministry of Education and Research Germany, speaker's honoraria from Bristol-Myers Squibb, Eli Lilly & Company, Novartis, Eisai, Boehringer Ingelheim and Servier and travel funds from Janssen-Cilag, Eli Lilly & Company, Novartis, AstraZeneca, Lundbeck and the SALUS–Institute for Trend-Research and Therapy Evaluation in Mental Health. M.N. received research grants from the Federal Ministry of Education and Research, Germany, the German Research Foundation, BioRad Laboratories, Siemens AG, Zeitschrift für Laboratoriumsmedizin, Bruker Daltronics, Abbott, Jurilab Kuopio, Roche Diagnostics, Instand and Becton Dickinson. H.V. received external research support via research grants from Hofmann La Roche, the Humboldt Foundation, the Federal Ministry of Education and Research (Germany) and the German Research Foundation. M.W. is on the following scientific advisory boards: Lilly, Araclon and Institut Catala de Neurociencies Aplicades, the Gulf War Veterans Illnesses Advisory Committee, VACO, Biogen Idec and Pfizer. M.W. received funding for consulting from Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, SA, Servier, Synarc, Pfizer and Janssen; for travel from NeuroVigil, CHRU–Hopital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, the University of California, San Diego–ADNI, Paris University, Institut Catala de Neurociencies Aplicades, the University of New Mexico School of Medicine, Ipsen, Clinical Trials on Alzheimer's Disease (CTAD), Pfizer, AD PD Meeting, Paul Sabatier University, Novartis and Tohoku University; and research support from: Merck, Avid, DoD, VA. M.W. received honoraria from PMDA/ the Japanese Ministry of Health, Labour, and Welfare, Tohoku University, Neuro Vigil, Insitut Catala de Neurociencies Aplicades. M.W. owns stock options for Synarc, Elan. Organizations contributing to the Foundation for the US NIH and thus to the National Institute on Aging (NIA)-funded Alzheimer's Disease Neuroimaging Initiative included Abbott, the Alzheimer's Association, the Alzheimer's Drug Discovery Foundation, Anonymous Foundation, AstraZeneca, Bayer Healthcare, BioClinica (ADNI 2), Bristol-Myers Squibb, the Cure Alzheimer's Fund, Eisai, Elan, Gene Network Sciences, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck, Novartis, Pfizer, Roche, Schering Plough, Synarc and Wyeth.
ADNI: The ADNI study was supported by the US NIH (U01 AG024904) and the Foundation for the NIH for genotype and phenotype data collection, the NIH (RC2 AG036535-01) for data analysis, the NIA (R01 AG019771-09) for additional data analysis and NCRAD (U24AG021886) for DNA used in part for the GWAS. Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (see URLs). As such, the investigators within ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. The ADNI sample wishes to acknowledge the investigators who contributed to the design and implementation of ADNI (see URLs). Data collection and sharing for this project were funded by ADNI (NIH grant U01 AG024904). ADNI is funded by the NIA, the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and through generous contributions from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck and Cocpany, Novartis AG, Pfizer, F. Hoffman–La Roche, Schering-Plough and Synarc, as well as from nonprofit partners at the Alzheimer's Association and the Alzheimer's Drug Discovery Foundation, with participation from the US Food and Drug Administration (FDA). Private sector contributions to ADNI are facilitated by the Foundation for the NIH (see URLs). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants (P30 AG010129 and K01 AG030514) and by the Dana Foundation. ADNI was launched in 2003 by the NIA, the NIBIB, the FDA, private pharmaceutical companies and nonprofit organizations as a 5-year public- private partnership. The primary goal of ADNI has been to test whether serial MRI), positron emission tomography (PET), other biological markers and clinical and neuropsychological assessments can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease. Determination of sensitive and specific markers of very early Alzheimer's disease progression is intended to aid researchers and clinicians in developing new treatments and monitoring their effectiveness, as well as lessening the time and cost of clinical trials. The Principal Investigator of this initiative is M.W. Weiner. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the United States and Canada. The initial goal of ADNI was to recruit 800 adults ages 55 to 90 to participate in the research—approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early Alzheimer's disease to be followed for 2 years. For up-to-date information, please visit the ADNI website (see URLs).
BIG: The BIG study wishes to acknowledge S. Kooijman for coordination of sample collection and A. Heister, M. Naber, R. Makkinje, M. Hakobjan and M. Steehouwer for genotyping. The BIG study was supported by a Biobanking and Biomolecular Resources Research Infrastructure Netherlands (BBMRI-NL) complementation grant for brain segmentation and the Netherlands Organisation for Scientific Research (NWO) Horizon Breakthrough grant (grant number 93511010 (to A.A.V.).
Bipolar Family Study: The Bipolar Family Study wishes to thank the Scottish Mental Health Research Network for research assistant support, the Brain Research Imaging Centre Edinburgh (see URLs), a center in the Scottish Funding Council Scottish Imaging Network–A Platform for Scientific Excellence (SINAPSE) Collaboration (see URLs), for image acquisition and the Wellcome Trust Clinical Research Facility for genotyping. Genotyping was supported by the National Alliance for Research on Schizophrenia and Depression (NARSAD) Independent Investigator Award (to A.M.M.), and data collection was supported by the Health Foundation Clinician Scientist Fellowship.
fBIRN: fBIRN wishes to acknowledge D.B. Keator for leading fBIRN neuroinformatics development, B.A. Mueller for image calibration and quality assurance and A. Belger, V.D. Calhoun, G.G. Brown, J.M. Ford, G.H. Glover, R. Kikinis, K. Lim, J. Laurriello, J. Bustillo, G. McCarthy, D.S. O'Leary, B. Rosen, A.W.T. and J.T. Voyvodic for their leadership contributions to fBIRN scanner and sequence calibration, tool development and data collection efforts. The fBIRN study was supported by the US NIH (U24 RR21992) for phenotypic data collection. Genotyping was performed with the support of the grant RBIN04SWHR to F.M. from the Italian Ministry of University and Research.
GOBS: The GOBS study was supported by the US NIH (MH0708143 and MH083824 to D.C.G., MH078111 and MH59490 to J.B., C06 RR13556 and C06 RR017515). P.K. was also supported by an NIH grant (EB006395).
IMAGEN: IMAGEN is funded by the European Commission Framework Programme 6 (FP-6) Integrated Project IMAGEN (PL037286), the European Commission Framework Programme 7 (FP-7) Project Alzheimer's Disease, Alcoholism, Memory, Schizophrenia (ADAMS), the FP-7 Innovative Medicine Initiative Project European Autism Interventions (AIMS), the UK Department of Health National Institute of Health Research (NIHR)–Biomedical Research Centre Mental Health program and the MRC programme grant Developmental Pathways into Adolescent Substance Abuse (93558).
ImaGene: ImaGene wishes to acknowledge J. Lee and J. Lane for processing the blood samples, The Easton Consortium for Alzheimer's Disease Drug Discovery and Biomarker Development and the Alzheimer's Disease Research Center (ADRC) funded by the NIA at the University of California, Los Angeles (AG16570).
LBC1936: We thank the participants in LBC1936. We thank C. Murray, A.J. Gow, S.E. Harris, M. Luciano, P. Redmond, E. Sandeman, I. Gerrish, J. Boyd-Ellison, N. Leslie, A. Howden and C. Scott for data collection and preparation. This project is funded by the Age UK's Disconnected Mind programme and also by Research Into Ageing (251 and 285). The entire genome association part of the study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/F019394/1). Analysis of brain images was funded by UK MRC grants (G1001401 and 8200). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Imaging was performed at the Brain Research Imaging Centre, Edinburgh, a center in the SINAPSE Collaboration. Funding from BBSRC, the Engineering and Physical Sciences Research Council (EPSRC), the Economic and Social Research Council (ESRC) and the MRC and Scottish Funding Council through the SINAPSE Collaboration is gratefully acknowledged. L.M.L. is the beneficiary of a postdoctoral grant from the AXA Research Fund.
MooDS: This work was funded by the German Federal Ministry of Education and Research (BMBF) in the National Genome Research Network (NGFN-plus) through the MooDs grant Molecular Causes of Major Mood Disorders and Schizophrenia (coordinator M.M.N.). Additional funding for genotyping was provided by a NARSAD Distinguished Investigator award to A.M.-L.
MPIP: The MPIP Munich Morphometry Sample comprises images acquired as part of the Munich Antidepressant Response Signature Study and the Recurrent Unipolar Depression (RUD) Case-Control Study performed at the MPIP and control subjects acquired at the Department of Psychiatry at the Ludwig-Maximilians-University. We wish to acknowledge A. Olynyik and radiographers R. Schirmer, E. Schreiter and R. Borschke for image acquisition and data preparation. We thank D.P. Auer for local study management in the initial phase of the RUD study. We are grateful to GlaxoSmithKline for providing the genotypes of the RUD Case-Control Sample. We thank the staff of the Center of Applied Genotyping (CAGT) for generating the genotypes of the MARS cohort. The study is supported by a grant from the Exzellenz-Stiftung of the Max Planck Society. This work has also been funded by the BMBF in the framework of the National Genome Research Network (NGFN) (FKZ 01GS0481).
NCNG: We would like to thank the personnel involved in recruitment and data collection and, in particular, P. Due-Tønnessen for clinical assessment of the MRI images. The NCNG study was supported by Research Council of Norway grants (154313/V50 and 177458/V50). The NCNG GWAS was financed by grants from the Bergen Research Foundation, the University of Bergen, the Research Council of Norway (FUGE; Psykisk Helse), Helse Vest Regionalt Helseforetak (RHF) and the Dr Einar Martens Fund.
NESDA-NTR: Funding was obtained from the NWO (MagW/ZonMW 904-61-090; 985-10-002; 904-61-193; 480-04-004; 400-05-717, Addiction-31160008; 911-09-032; SPI 56-464-14192 and Geestkracht Program, 10-000-1002), the Center for Medical Systems Biology (CMSB; NWO Genomics), NBIC/BioAssist/RK/2008.024, BBMRI-NL, Biobanking and Biomolecular Resources Research Infrastructure, the VU University, the EMGO Institute for Health and Care Research and Neuroscience Campus Amsterdam, the European Science Foundation (EU/QLRT-2001-01254), the European Community's FP7 (HEALTH-F4-2007-201413), the European Science Council (ERC) Genetics of Mental Illness (230374), Rutgers University Cell and DNA Repository (cooperative agreement NIMH U24 MH068457-06), the US NIH (R01D0042157-01A) and the Genetic Association Information Network (a public-private partnership between the NIH and Pfizer, Affymetrix and Abbott Laboratories).
NIMH-IRP: This study was supported by funding from the Intramural Research Program of the National Institute of Mental Health (NIMH) from the NIH and the US Department of Health and Human Services (K99 MH085098 to G.L., 1ZIA MH002810 to F.J.M. and 1ZIA MH002790 to W.C.D.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government.
QTIM: We are extremely grateful to the twins for their participation, the radiographers at the Centre for Advanced Imaging at the University of Queensland for image acquisition and the many research assistants and support staff at the Queensland Institute of Medical Research for twin recruitment and daily management, and we especially thank K. Johnson for MRI scanning and processing, A. Henders for DNA processing and preparation and S. Gordon for quality control and management of the genotypes. Phenotyping was funded by the US National Institute of Child Health and Human Development (R01 HD050735) and the Australian National Health and Medical Research Council (NHMRC) (project grant 496682). Genotyping was funded by the NHMRC (Medical Bioinformatics Genomics Proteomics Program, 389891). G.M. was supported by an NHMRC Fellowship (613667), and G.Z. was supported by Australian Research Council (ARC) Future Fellowship (FT0991634). S.E.M. is funded by an ARC Future Fellowship (FT110100548). J.L.S. was supported by the Achievement Rewards for College Scientists foundation and the US NIMH (F31 MH087061). D.P.H. is partially supported by a National Science Foundation (NSF) Graduate Research Fellowhip Program (GRFP) grant (DGE-0707424). P.T. was also supported by the NIH (grants U01 AG024904, AG040060, EB008432, P41 RR013642, HD050735, AG036535, AG020098 and EB008281).
SYS: The Saguenay Youth Study Group wishes to thank the following individuals for their contribution in acquiring and analyzing the data: N. Arbour, M.-È. Bouchard, A. Houde, A. Gauthier and H. Simard for the recruitment and assessment of participating families, M. Bérubé, S. Masson, S. Castonguay and M.-J. Morin for MRI acquisition and E. Ding and N. Qiu for MR data management. We thank J. Mathieu for the medical follow up of participants in whom we detected any medically relevant abnormalities. We are grateful to all families for participating in the study. The Saguenay Youth Study Group is supported by the Canadian Institutes of Health Research, the Heart and Stroke Foundation of Quebec and the Canadian Foundation for Innovation. For more information, please see the study website (see URLs).
SHIP: The Study of Health in Pomerania (SHIP) is supported by the German Federal Ministry of Education and Research (grants 01ZZ9603, 01ZZ0103 and 01ZZ0403) and the German Research Foundation (DFG; GR 1912/5-1). Genome-wide data and MRI scans were supported by the Federal Ministry of Education and Research (grant 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Center of Knowledge Interchange program of the Siemens AG. We thank all staff members and participants of the SHIP study, as well as all of the genotyping staff for generating the SHIP SNP data set. The genetic data analysis workflow was created using the Software InforSense. Genetic data were stored using the database Caché (InterSystems).
SHIP-TREND: The authors from SHIP are grateful to M. Stanke for the opportunity to use his Server Cluster for SNP Imputation. This cohort is part of the Community Medicine Research net (CMR) of the University of Greifswald, which is funded by the German Federal Ministry of Education and Research and the German Ministry of Cultural Affairs, as well as by the Social Ministry of the Federal State of Mecklenburg–West Pomerania. CMR encompasses several research projects that share data from the population-based Study of Health in Pomerania (SHIP; see URLs). The work is also supported by the German Research Foundation (DFG; GR 1912/5-1) and the Greifswald Approach to Individualized Medicine (GANI_MED) network funded by the Federal Ministry of Education and Research (grant 03IS2061A). Genome-wide data and MRI scans were supported by the Federal Ministry of Education and Research (grant 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. The University of Greifswald is a member of the Center of Knowledge Interchange program of the Siemens AG.
Superstruct: We thank the investigators and participants who contributed to the brain genomics data collection for Superstruct at Massachusetts General Hospital and Harvard University, with funding from the Simons Foundation, the Howard Hughes Medical Institute and the US NIH (grant MH079799).
TOP: We thank the study participants of TOP and the personnel involved in data collection and logistics, especially T.D. Bjella. This work was supported by the Oslo University Hospital–Ullevål, the Eastern Norway Health Authority (2004-123), the Research Council of Norway (167153/V50, 163070/V50 and 183782/V50), and by Eli Lilly & Company (who covered part of the genotyping costs).
TCD: We wish to express our sincere thanks to all participants and to clinical staff who facilitated patients' involvement. In particular, we acknowledge colleagues from the Trinity College Institute of Neuroscience A. Bodke, J. McGrath, F. Newell, H. Garavan, and J. O'Doherty for their support in sample collection. Collection and analysis of these samples were funded by the Wellcome Trust (072894/z/03/z-Gill) and the Science Foundation Ireland (08/IN.1/B1916_Corvin).
EPIGEN: Work from the London Cohort was supported by research grants from the Wellcome Trust (grant 084730 to S.M.S.), University College London (UCL)/University College London Hospitals (UCLH) Comprehensive Biomedical Research Centre/Specialist Biomedical Research Centres (CBRC/SBRC) (grant 114 to S.M.S.), the European Union Marie Curie Reintegration (to M. Matarin and S.M.S.), the UK NIHR (08-08-SCC), the Comprehensive Local Research Network (CLRN) Flexibility and Sustainability Funding (FSF) (grant CEL1300 to S.M.S.), The Big Lottery Fund, the Wolfson Trust and the Epilepsy Society. This work was undertaken at UCLH/UCL, which received a proportion of funding from the UK Department of Health's NIHR Biomedical Research Centres funding scheme. Work from the Royal College of Surgeons in Ireland was supported by research grants from the Science Foundation Ireland (Research Frontiers Programme award 08/RFP/GEN1538) and Brainwave–the Irish Epilepsy Association. The collection of Belgian subjects was supported by the Fonds National de la Recherche Scientifique (grant FC 63574 / 3.4.620.06 F) and the Fonds Erasme pour la Recherche Médicale at the Université Libre de Bruxelles.
UCL Institute of Neurology Control Brain Tissue Collection: Funding was provided by the UK MRC (grant G0901254), the MRC Sudden Death Brain and Tissue Bank and the Sun Health Research Institute Brain Bank.
UMCU: The UMCU study was supported by the Netherlands Organization for Health Research and Development ZonMw (917.46.370 to H.E.H.) and the US NIMH (MH078075 to R.A.O.).
Author information
Authors and Affiliations
Consortia
Contributions
The ENIGMA support group designed the project, established the consortium, determined the analysis and quality control procedures, offered analytical support and performed and coordinated cross-site and replication analyses. This group included J.L.S., S.E.M., A.A.V., D.P.H., M.J.W., B.F., N.G.M. and P.M.T. The imaging protocols group determined and refined protocols for computing brain measures from the MRI scans and helped sites implement them as needed. This group included J.L.S., R.T., A.M.W., T.E.N., M.J. and M. Rijpkema. The genetics protocols group created analysis methods for imputation, quality control and association testing of genome-wide data and helped to ensure that protocols were implemented consistently across all sites. This group included S.E.M., J.L.S., A.A.V. and D.P.H. The meta-analysis was carried out by the meta-analysis group, consisting of S.E.M., R.E.S., J.L.S., D.P.H., A.A.V., M.J.W., N.G.M., B.F. and P.M.T. The first draft was written by J.L.S., S.E.M., A.A.V., D.P.H., M.J.W., B.F., N.G.M. and P.M.T. Local image processing, involving statistical analysis and analysis of the data, was performed by J.L.S., A.M.W., D.P.H., R.B., Ø.B., M.M.C., O.G., M. Hollinshead, A.J.H., S.M.M., A.C.N., M. Rijpkema, N.A.R., M.C.V.H., T.G.M.v.E., S.W., D.G.B., S.L.R., J.L.R., M.-J.v.T., S.E., P.T.F., P.K., J.L.L., R.M., G.B.P., J. Savitz, H.G.S., K.S., A.W.T., M.V.d.H., N.J.v.d.W., N.E.M.V.H., H.W., A.M.D., C.R.J., D.J.V., E.J.C.d.G., G.I.d.Z., T.E., G.F., P.H., H.E.H.P., K.L.M., A.J.S., L.S., J.B., D.C.G., K.N., E.L., A.M.-L., P.G.S., L.G.A., K.S.H., T.P., M.D., R.P., N.H., K.W., I.A., Ø.B., A.M.D., D.H., M.C., S.A., N.D., C. Depondt, M. Pandolfo, E.J.R., D.M.C., J.C.R., J.R., J.T., R.T., C.L., S.M., A.H., C.D.W., N.J., D.J.H., L.T.W. and M. Hoogman. Local genetics processing, involving statistical analysis and analysis of the data, was performed by J.L.S., S.E.M., A.A.V., A.M.W., D.P.H., M.B., A.A.B., A. Christoforou, G. Davies, J.-J.H., L.M.L., G.L., P.H.L., D.C.L., X.L., M. Mattingsdal, K.N., E. Strengman, K.v.E., T.G.M.v.E., S.W., S.K., L.A., R.M.C., M.A.C., J.E.C., R.D., T.D.D., N.B.F., H.H.H.G., M.P.J., J.W.K., M. Mattheisen, E.K.M., T.W.M., M.M.N., M. Rietschel, V.M.S., A.W.T., J.A.V., S.C., S.D., T.M.F., P.H., S.L.H., G.W.M., O.A.A., H.G.B., R.A.O., B.W.P., A.J.S., L.S., J.B., D.C.G., M.J.W., N.G.M., A.L., E.B.B., C.W., B.P., B.M.-M., G.C., Z.P., G.H., M.N., A.T., D.K., M. Matarin, S.M.S., G.L.C., N.K.H., M.E.R., D.W.M., C.O., A. Corvin, M.G., J.F., J.C.R., A.R., M. Ryten, D.T., N.S., C.S., R.W., J. Hardy, M.E.W. and M.A.A.d.A. Local study oversight and management, involving joint supervision of research, contribution of reagents, materials and/or analysis tools, was carried out by R.L.B., R.D., P.T.F., R.S.K., I.M., R.L.O., I.R., I.A., W.C.D., P.H., F.M., A.M.-L., D.J.P., S.G.P., J.M.S., M.W.W., O.A.A., M.E.B., H.G.B., E.J.C.d.G., I.J.D., G.I.d.Z., T.E., G.F., H.E.H.P., F.J.M., K.L.M., R.A.O., T.P., Z.P., B.W.P., A.J.S., L.S., J.W.S., J.M.W., J.B., D.C.G., M.J.W., B.F., P.M.T., A.M.M., J. Hall, M. Papmeyer, E. Sprooten, J. Sussmann, S.M.L., J.B.P., L.G.A., G.C., D.R., E.M., G.S., K.S.H., P.G.S., E.B.B., D.I.B., H.J.G., H.V., K.A., C.M., G. Donohoe, F.H., A.V.S., V.G., C.T., M.W.V., L.J.L., C. DeCarli, S.S., J.C.B., M.A.I., A.A. and J. Hardy.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–44 and Supplementary Tables 1–27 (PDF 25711 kb)
Rights and permissions
About this article
Cite this article
Stein, J., Medland, S., Vasquez, A. et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44, 552–561 (2012). https://doi.org/10.1038/ng.2250
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2250
This article is cited by
-
Pathogenesis of Depression in Alzheimer’s Disease
Neurochemical Research (2024)
-
Posterior hippocampal CA2/3 volume is associated with autobiographical memory recall ability in lower performing individuals
Scientific Reports (2023)
-
Leveraging molecular quantitative trait loci to comprehend complex diseases/traits from the omics perspective
Human Genetics (2023)
-
Genetic and Environmental Contributions to Subcortical Gray Matter Microstructure and Volume in the Developing Brain
Behavior Genetics (2023)
-
Identifying genes associated with brain volumetric differences through tissue specific transcriptomic inference from GWAS summary data
BMC Bioinformatics (2022)