Abstract
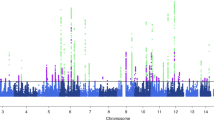
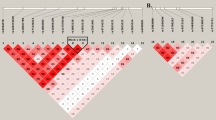
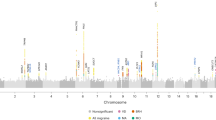
Migraine without aura is the most common form of migraine, characterized by recurrent disabling headache and associated autonomic symptoms. To identify common genetic variants associated with this migraine type, we analyzed genome-wide association data of 2,326 clinic-based German and Dutch individuals with migraine without aura and 4,580 population-matched controls. We selected SNPs from 12 loci with 2 or more SNPs associated with P values of <1 × 10−5 for replication testing in 2,508 individuals with migraine without aura and 2,652 controls. SNPs at two of these loci showed convincing replication: at 1q22 (in MEF2D; replication P = 4.9 × 10−4; combined P = 7.06 × 10−11) and at 3p24 (near TGFBR2; replication P = 1.0 × 10−4; combined P = 1.17 × 10−9). In addition, SNPs at the PHACTR1 and ASTN2 loci showed suggestive evidence of replication (P = 0.01; combined P = 3.20 × 10−8 and P = 0.02; combined P = 3.86 × 10−8, respectively). We also replicated associations at two previously reported migraine loci in or near TRPM8 and LRP1. This study identifies the first susceptibility loci for migraine without aura, thereby expanding our knowledge of this debilitating neurological disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Launer, L.J., Terwindt, G.M. & Ferrari, M.D. The prevalence and characteristics of migraine in a population-based cohort: the GEM study. Neurology 53, 537–542 (1999).
Stovner, L.J., Zwart, J.A., Hagen, K., Terwindt, G.M. & Pascual, J. Epidemiology of headache in Europe. Eur. J. Neurol. 13, 333–345 (2006).
Stovner, L. et al. The global burden of headache: a documentation of headache prevalence and disability worldwide. Cephalalgia 27, 193–210 (2007).
Olesen, J., Lekander, I., Andlin-Sobocki, P. & Jönsson, B. Funding of headache research in Europe. Cephalalgia 27, 995–999 (2007).
Anttila, V. et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat. Genet. 42, 869–873 (2010).
Chasman, D.I. et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat. Genet. 43, 695–698 (2011).
Dimas, A.S. et al. Common regulatory variation impacts gene expression in a cell type–dependent manner. Science 325, 1246–1250 (2009).
Nica, A.C. et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 7, e1002003 (2011).
Lin, X., Shah, S. & Bulleit, R.F. The expression of MEF2 genes is implicated in CNS neuronal differentiation. Brain Res. Mol. Brain Res. 42, 307–316 (1996).
Flavell, S.W. et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 (2006).
Aurora, S.K. & Wilkinson, F. The brain is hyperexcitable in migraine. Cephalalgia 27, 1442–1453 (2007).
Ferrari, M.D., Odink, J., Bos, K.D., Malessy, M.J. & Bruyn, G.W. Neuroexcitatory plasma amino acids are elevated in migraine. Neurology 40, 1582–1586 (1990).
Martínez, F., Castillo, J., Rodríguez, J.R., Leira, R. & Noya, M. Neuroexcitatory amino acid levels in plasma and cerebrospinal fluid during migraine attacks. Cephalalgia 13, 89–93 (1993).
Tottene, A. et al. Enhanced excitatory transmission at cortical synapses as the basis for facilitated spreading depression in Cav2.1 knockin migraine mice. Neuron 61, 762–773 (2009).
Flavell, S.W. et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity–dependent polyadenylation site selection. Neuron 60, 1022–1038 (2008).
Morrow, E.M. et al. Identifying autism loci and genes by tracing recent shared ancestry. Science 321, 218–223 (2008).
Pfeiffer, B.E. et al. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron 66, 191–197 (2010).
Schytz, H.W. et al. PACAP38 induces migraine-like attacks in patients with migraine without aura. Brain 132, 16–25 (2009).
Markovics, A. et al. Pituitary adenylate cyclase–activating polypeptide plays a key role in nitroglycerol-induced trigeminovascular activation in mice. Neurobiol. Dis. 45, 633–644 (2012).
Goadsby, P.J., Lipton, R.B. & Ferrari, M.D. Migraine—current understanding and treatment. N. Engl. J. Med. 346, 257–270 (2002).
Lin, H.Y., Wang, X.F., Ng-Eaton, E., Weinberg, R.A. & Lodish, H.F. Expression cloning of the TGF-β type 2 receptor, a functional transmembrane serine/threonine kinase. Cell 68, 775–785 (1992).
Law, C. et al. Clinical features in a family with an R460H mutation in transforming growth factor β receptor 2 gene. J. Med. Genet. 43, 908–916 (2006).
Rist, P.M., Diener, H.C., Kurth, T. & Schürks, M. Migraine, migraine aura, and cervical artery dissection: a systematic review and meta-analysis. Cephalalgia 31, 886–896 (2011).
Allen, P.B., Greenfield, A.T., Svenningsson, P., Haspeslagh, D.C. & Greengard, P. Phactrs 1–4: a family of protein phosphatise 1 and actin regulatory proteins. Proc. Natl. Acad. Sci. USA 101, 7187–7192 (2004).
Greengard, P., Allen, P.B. & Nairn, A.C. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron 23, 435–447 (1999).
Jarray, R. et al. Depletion of the novel protein PHACTR-1 from human endothelial cells abolishes tube formation and induces cell death receptor apoptosis. Biochimie 93, 1668–1675 (2011).
Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
Tietjen, G.E. Migraine as a systemic vasculopathy. Cephalalgia 29, 987–996 (2009).
Wilson, P.M., Fryer, R.H., Fang, Y. & Hatten, M.E. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J. Neurosci. 30, 8529–8540 (2010).
DaSilva, A.F., Granziera, C., Snyder, J. & Hadjikhani, N. Thickening in the somatosensory cortex of patients with migraine. Neurology 69, 1990–1995 (2007).
Kruit, M.C. et al. Migraine as a risk factor for subclinical brain lesions. J. Am. Med. Assoc. 291, 427–434 (2004).
Proudfoot, C.J. et al. Analgesia mediated by the TRPM8 cold receptor in chronic neuropathic pain. Curr. Biol. 16, 1591–1605 (2006).
Caspani, O., Zurborg, S., Labuz, D. & Heppenstall, P.A. The contribution of TRPM8 and TRPA1 channels to cold allodynia and neuropathic pain. PLoS ONE 4, e7383 (2009).
d'Agostino, V.C., Francia, E., Licursi, V. & Cerbo, R. Clinical and personality features of allodynic migraine. Neurol. Sci. 31 (suppl. 1), S159–S161 (2010).
Lillis, A.P., van Duyn, L.B., Murphy-Ullrich, J.E. & Strickland, D.K. LDL receptor–related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol. Rev. 88, 887–918 (2008).
May, P. et al. Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol. Cell. Biol. 24, 8872–8883 (2004).
Russell, M.B., Rasmussen, B.K., Fenger, K. & Olesen, J. Migraine without aura and migraine with aura are distinct clinical entities: a study of four hundred and eighty-four male and female migraineurs from the general population. Cephalalgia 16, 239–245 (1996).
Kallela, M., Wessman, M., Havanka, H., Palotie, A. & Färkkilä, M. Familial migraine with and without aura: clinical characteristics and co-occurrence. Eur. J. Neurol. 8, 441–449 (2001).
Frazer, K.A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Teo, Y.Y. et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics 23, 2741–2746 (2007).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Altshuler, D.M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Mägi, R. & Morris, A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 11, 288 (2010).
Cochran, W.G. The combination of estimates from different experiments. Biometrics 10, 101–129 (1954).
Higgins, J.P. & Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Ioannidis, J.P., Patsopoulos, N.A. & Evangelou, E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS ONE 2, e841 (2007).
DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188 (1986).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population- based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Stranger, B.E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Acknowledgements
We wish to thank all individuals in the respective cohorts for their generous participation. This work was supported by the German Federal Ministry of Education and Research (BMBF) (grant 01GS08121 to M.D., along with support to E.W. in the context of the German National Genome Research Network, (NGFN-2 and NGFN-plus) for the Heinz Nixdorf Recall Study); the Spanish Ministry of Science and Innovation (grant SAF2009-13182-C03 to A.M. and B.C.); the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) (grants 2009SGR78 and 2009SGR971 to A.M. and B.C., respectively); an unrestricted grant of the Vascular Dementia Research Foundation (to M.D.); the Netherlands Organization for Health Research and Development (ZonMw) (90700217) and VIDI (ZonMw) (91711319 to G.M.T.); the Netherlands Organisation for Scientific Research (NWO) VICI (918.56.602) and Spinoza (2009) grants (to M.D.F.); the Center for Medical Systems Biology (CMSB) established in the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NGI/NWO) (project 050-060-409 to C.M.v.D., R.R.F., M.D.F. and A.M.J.M.v.d.M. and 050-060-810 to C.M.v.D.) and the generation and management of GWAS genotype data for the Rotterdam Study (175.010.2005.011 and 911-03-012) (funded by the Erasmus Medical Center, Erasmus University Rotterdam and the Ministries of Education, Culture and Science, Health, Welfare and Sports), as well as the NGI-sponsored Netherlands Consortium for Healthy Aging (NCHA) and the Research Institute for Diseases in the Elderly (014-93-015; RIDE2); the German Federal Ministry of Education and Research (BMBF) within the framework of the National Genome Research Network (NGFN-Plus; grants 01GS08120 and 01GS1103 to C.K.); the German Federal Ministry of Education and Research and the State of Bavaria, supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ) for the KORA research platform, which was initiated by the Helmholtz Center Munich, German Research Center for Environmental Health; the National Health and Medical Research Council (NHMRC) Research Fellowship (613674) and the Australian Research Council (ARC) Future Fellowship (FT0991022) schemes (to D.R.N.); the Wellcome Trust (grant 098051 to A.P.); the Academy of Finland (grant 251704 to A.P. and 139795 to M.W.); the Academy of Finland, Center of Excellence in Complex Disease Genetics (grants 213506 and 129680 to A.P. and J.K.); the South-Eastern Norway Regional Health Authority (2010075 and 2011083 to B.W. and J.-A.Z.), the Unger-Vetlesen Medical Fund (to B.W.) and the Ullevaal fund (to B.W.); the European Community's Seventh Framework Programme (FP7/2007-2013), the ENGAGE Consortium, (grant agreement HEALTH-F4-2007- 201413); EU/SYNSYS–Synaptic Systems (grant 242167 to A.P.); the Sigrid Juselius Foundation, Finland (to A.P.); the Folkhälsan Research Foundation, Helsinki (to M.W.); and the Helsinki University Central Hospital (to M.K., V. Artto and M.F.).
Author information
Authors and Affiliations
Consortia
Contributions
Funding was obtained by M.D., M.D.F., A.P., A.M.J.M.v.d.M., C.K. and C.M.v.D. Overall study design was guided by T.F., V. Anttila, B.d.V., R.M., D.R.N., J.-A.Z., C.K., A.P., M.D. and A.M.J.M.v.d.M. Cohorts were supervised and phenotyped by T.F., M.K., G.M.T., P.P.-R., B.W., W.P.J.v.O., V. Artto, U.T., J.F.-M., M.A.L., M.A.K., J.S., O.R., T.L., M.V.-P., H.G., E.W., C.S., A.G.U., A. Heinze, A. Hoffman., E.T., C.M.v.D., J.K., B.C., T.M., J.-A.Z., M.F. and A.M. Analysis and genotyping were performed by V. Anttila, B.d.V., R.M., B.W., D.R.N., E.H., A.G.U., F.R., M.W., T.M. and B.M.-M. The manuscript was written by T.F., V. Anttila, B.d.V., D.R.N., B.C., M.W., R.R.F., J.-A.Z., C.K., A.P., M.D. and A.M.J.M.v.d.M. All authors participated in critical review of the manuscript for intellectual content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–3 and Supplementary Note (PDF 259 kb)
Rights and permissions
About this article
Cite this article
Freilinger, T., Anttila, V., de Vries, B. et al. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet 44, 777–782 (2012). https://doi.org/10.1038/ng.2307
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2307
This article is cited by
-
Genetics of migraine: where are we now?
The Journal of Headache and Pain (2023)
-
Current Update on Categorization of Migraine Subtypes on the Basis of Genetic Variation: a Systematic Review
Molecular Neurobiology (2023)
-
Migraine: from pathophysiology to treatment
Journal of Neurology (2023)
-
Advancements in the Genetics of Spontaneous Coronary Artery Dissection
Current Cardiology Reports (2023)
-
A Meta-Analysis of the Genome-Wide Association Studies on Two Genetically Correlated Phenotypes Suggests Four New Risk Loci for Headaches
Phenomics (2023)