Abstract
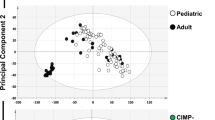
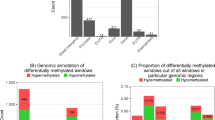
We have extensively characterized the DNA methylomes of 139 patients with chronic lymphocytic leukemia (CLL) with mutated or unmutated IGHV and of several mature B-cell subpopulations through the use of whole-genome bisulfite sequencing and high-density microarrays. The two molecular subtypes of CLL have differing DNA methylomes that seem to represent epigenetic imprints from distinct normal B-cell subpopulations. DNA hypomethylation in the gene body, targeting mostly enhancer sites, was the most frequent difference between naive and memory B cells and between the two molecular subtypes of CLL and normal B cells. Although DNA methylation and gene expression were poorly correlated, we identified gene-body CpG dinucleotides whose methylation was positively or negatively associated with expression. We have also recognized a DNA methylation signature that distinguishes new clinico-biological subtypes of CLL. We propose an epigenomic scenario in which differential methylation in the gene body may have functional and clinical implications in leukemogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rozman, C. & Montserrat, E. Chronic lymphocytic leukemia. N. Engl. J. Med. 333, 1052–1057 (1995).
Zenz, T., Mertens, D., Kuppers, R., Dohner, H. & Stilgenbauer, S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer 10, 37–50 (2010).
Chiorazzi, N. & Ferrarini, M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood 117, 1781–1791 (2011).
Baylin, S.B. & Jones, P.A. A decade of exploring the cancer epigenome—biological and translational implications. Nat. Rev. Cancer 11, 726–734 (2011).
Cedar, H. & Bergman, Y. Epigenetics of haematopoietic cell development. Nat. Rev. Immunol. 11, 478–488 (2011).
Deaton, A.M. et al. Cell type–specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 21, 1074–1086 (2011).
Rodríguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–339 (2011).
Kanduri, M. et al. Differential genome-wide array–based methylation profiles in prognostic subsets of chronic lymphocytic leukemia. Blood 115, 296–305 (2010).
Rahmatpanah, F.B. et al. Large-scale analysis of DNA methylation in chronic lymphocytic leukemia. Epigenomics 1, 39–61 (2009).
Tong, W.G. et al. Genome-wide DNA methylation profiling of chronic lymphocytic leukemia allows identification of epigenetically repressed molecular pathways with clinical impact. Epigenetics 5, 499–508 (2010).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Berman, B.P. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina–associated domains. Nat. Genet. 44, 40–46 (2012).
Shaknovich, R. et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 118, 3559–3569 (2011).
Bibikova, M. et al. High-density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011).
Klein, U. et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194, 1625–1638 (2001).
Rosenwald, A. et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J. Exp. Med. 194, 1639–1648 (2001).
Kanber, D. et al. The human retinoblastoma gene is imprinted. PLoS Genet. 5, e1000790 (2009).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Maunakea, A.K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Hansen, K.D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Ohm, J.E. et al. A stem cell–like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39, 237–242 (2007).
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236 (2007).
Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007).
Shukla, S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011).
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011).
Quesada, V. et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 44, 47–52 (2012).
Wang, L. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Chan, T.A. et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. 5, e114 (2008).
Puente, X.S. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011).
Jones, P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012).
Adams, D. et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 30, 224–226 (2012).
The International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 993–998 (2010).
Bibikova, M. et al. Genome-wide DNA methylation profiling using Infinium assay. Epigenomics 1, 177–200 (2009).
Du, P., Kibbe, W.A. & Lin, S.M. Lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Marco-Sola, S., Sammeth, M., Guigó, R. & Ribeca, P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat. Methods (in the press).
Gautier, L., Cope, L., Bolstad, B.M. & Irizarry, R.A. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004).
Simpson, T.I., Armstrong, J.D. & Jarman, A.P. Merged consensus clustering to assess and improve class discovery with microarray data. BMC Bioinformatics 11, 590 (2010).
Acknowledgements
We thank M. Seifert for advice about isolating normal mature B-cell subpopulations and C. Rozman, J. Valcárcel, R. Küppers and R. Siebert for their comments on the manuscript. We are grateful to S. Guijarro, S. Martín, C. Capdevila, M. Sánchez and L. Plà for excellent technical assistance; to M. Lozano for helping us obtain normal peripheral blood samples; and to N. Villahoz and C. Muro for excellent work in the coordination of the CLL Spanish Consortium. We are indebted to the Hospital Clínic de Barcelona–IDIBAPS Biobank-Tumor Bank and Hematopathology Collection for the sample procurement. We are also very grateful to the patients with CLL who have participated in this study. This work was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the Instituto de Salud Carlos III (ISCIII) (to E.C. and C.L.-O.) and the Red Temática de Investigación del Cáncer (RTICC) of the ISCIII (to E.C.), project SAF2009-08663 (to J.I.M.-S.) and project MCyT-BIO2007-666855 (to A.V.), as well as the European Union's Seventh Framework Programme through the Blueprint Consortium (grant agreement 282510) (to E.C. and I.G.) and the Botín Foundation (to C.L.-O.). J.I.M.-S. is supported by a Ramon y Cajal contract of the MINECO, M.K. by the Agència de Gestió d'Ajuts Universitaris i de Recerca (Generalitat de Catalunya), A.C.Q. by the Portuguese Fundação para a Ciência e a Tecnologia, S.B.-S. by an EMBO Long-Term Fellowship and S.E. by La Caixa.
Author information
Authors and Affiliations
Contributions
M.K. and A.C.Q. purified normal B cells, analyzed DNA methylation and gene expression arrays and integrated the data. S.H., M. Rubio, R.R. and D.G.P. processed and analyzed WGBS data. M. Bibikova, V.H., B.K. and J.-B.F. performed Infinium 450k microarray experiments and primary data analysis. A.N. performed mutational analysis of IGHV genes. G. Clot, M.K., A.C.Q., G. Castellano, A.M.-T. and N.V. performed statistical analysis. A.M.-T., N.V., M.A., M. Rozman, E.C. and A.L.-G. reviewed the pathologic and clinical data and confirmed diagnoses. I.B.-H., M. Bayes and M.G. performed WGBS library preparation and sequencing. M.P. and P.J. performed gene expression microarray experiments. S.B.-S., P.P., A.C.Q., L.H. and M.L.-G. performed gene expression and/or alternative splicing studies. S.B. performed copy-number analyses. G. Castellano, D.R., S.E. and A.V. functionally characterized differentially methylated regions. L.C., D.C., M.P. and M.A. coordinated or performed sample preparation and quality control. V.Q., X.S.P. and M.K. integrated DNA methylation and gene mutation data. M. Rubio, R.R., J.L.G., M.O., D.G.P. and A.V. were in charge of data management. I.G. coordinated sequencing efforts and performed primary data analysis. C.L.-O., E.C. and J.I.M.-S. conceived of the study. J.I.M.-S. led the experiments and wrote the paper with assistance from M.K., S.H., A.C.Q., C.L.-O. and E.C.
Corresponding author
Ethics declarations
Competing interests
M.Bibikova., V.H., B.K. and J.-B.F. are employees of Illumina, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–21 and Supplementary Tables 1–4, 9,10,12–14,16–18 (PDF 2662 kb)
Supplementary Table 5
Annotation of differentially methylated CpGs between CLL subgroups (XLS 517 kb)
Supplementary Table 6
Annotation of differentially methylated CpGs between U-CLL and CD5+NBC/NBC (XLS 4769 kb)
Supplementary Table 7
Annotation of differentially methylated CpGs between M-CLL and MBC (XLS 762 kb)
Supplementary Table 8
Lists of genes differentially methylated in distinct parts of the gene length in CLLs vs. controls by microarrays (XLS 297 kb)
Supplementary Table 11
Genes and CpGs showing a significant correlation between expression and DNA methylation levels (XLS 607 kb)
Supplementary Table 15
Differentially methylated CpGs located in the proximity of alternative splicing regions (XLS 146 kb)
Rights and permissions
About this article
Cite this article
Kulis, M., Heath, S., Bibikova, M. et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 44, 1236–1242 (2012). https://doi.org/10.1038/ng.2443
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2443
This article is cited by
-
DDM1-mediated gene body DNA methylation is associated with inducible activation of defense-related genes in Arabidopsis
Genome Biology (2023)
-
Genome-wide DNA methylation and transcriptome analyses reveal the key gene for wool type variation in sheep
Journal of Animal Science and Biotechnology (2023)
-
Molecular characterization of Richter syndrome identifies de novo diffuse large B-cell lymphomas with poor prognosis
Nature Communications (2023)
-
Genomic landscape of mature B-cell non-Hodgkin lymphomas — an appraisal from lymphomagenesis to drug resistance
Journal of the Egyptian National Cancer Institute (2022)
-
Aberrant epigenetic and transcriptional events associated with breast cancer risk
Clinical Epigenetics (2022)