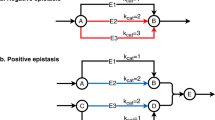
Abstract
Epistasis refers to the interaction between genes. Although high-throughput epistasis data from model organisms are being generated and used to construct genetic networks1,2,3, the extent to which genetic epistasis reflects biologically meaningful interactions remains unclear4,5,6. We have addressed this question through in silico mapping of positive and negative epistatic interactions amongst biochemical reactions within the metabolic networks of Escherichia coli and Saccharomyces cerevisiae using flux balance analysis. We found that negative epistasis occurs mainly between nonessential reactions with overlapping functions, whereas positive epistasis usually involves essential reactions, is highly abundant and, unexpectedly, often occurs between reactions without overlapping functions. We offer mechanistic explanations of these findings and experimentally validate them for 61 S. cerevisiae gene pairs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tischler, J., Lehner, B. & Fraser, A.G. Evolutionary plasticity of genetic interaction networks. Nat. Genet. 40, 390–391 (2008).
Boone, C., Bussey, H. & Andrews, B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8, 437–449 (2007).
Roguev, A. et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322, 405–410 (2008).
Moore, J.H. & Williams, S.M. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. Bioessays 27, 637–646 (2005).
Cordell, H.J. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 11, 2463–2468 (2002).
Phillips, P.C. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867 (2008).
Bateson, W. Mendel's Principles of Heredity (Cambridge University Press, Cambridge, UK, 1909).
Fisher, R.A. The correlations between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52, 399–433 (1918).
Hartman, J.L., Garvik, B. & Hartwell, L. Principles for the buffering of genetic variation. Science 291, 1001–1004 (2001).
Coyne, J.A. Genetics and speciation. Nature 355, 511–515 (1992).
Kondrashov, A.S. Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440 (1988).
Barton, N.H. & Charlesworth, B. Why sex and recombination? Science 281, 1986–1990 (1998).
Kondrashov, A.S. & Crow, J.F. Haploidy or diploidy: which is better? Nature 351, 314–315 (1991).
Crow, J.F. & Kimura, M. Efficiency of truncation selection. Proc. Natl. Acad. Sci. USA 76, 396–399 (1979).
Jasnos, L. & Korona, R. Epistatic buffering of fitness loss in yeast double deletion strains. Nat. Genet. 39, 550–554 (2007).
Yeh, P., Tschumi, A.I. & Kishony, R. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38, 489–494 (2006).
Segrè, D., Deluna, A., Church, G.M. & Kishony, R. Modular epistasis in yeast metabolism. Nat. Genet. 37, 77–83 (2005).
Price, N.D., Reed, J.L. & Palsson, B.O. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2, 886–897 (2004).
Papp, B., Pal, C. & Hurst, L.D. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429, 661–664 (2004).
Ibarra, R.U., Edwards, J.S. & Palsson, B.O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (2002).
Harrison, R., Papp, B., Pal, C., Oliver, S.G. & Delneri, D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. USA 104, 2307–2312 (2007).
Deutscher, D., Meilijson, I., Kupiec, M. & Ruppin, E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat. Genet. 38, 993–998 (2006).
Edwards, J.S., Ibarra, R.U. & Palsson, B.O. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19, 125–130 (2001).
Reed, J.L., Vo, T.D., Schilling, C.H. & Palsson, B.O. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4, R54 (2003).
Szathmáry, E. Do deleterious mutations act synergistically? Metabolic control theory provides a partial answer. Genetics 133, 127–132 (1993).
Deutschbauer, A.M. et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169, 1915–1925 (2005).
Kishony, R. & Leibler, S. Environmental stresses can alleviate the average deleterious effect of mutations. J. Biol. 2, 14 (2003).
Breslow, D.K. et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711–718 (2008).
Elena, S.F. & Lenski, R.E. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390, 395–398 (1997).
Liao, B.Y. & Zhang, J. Mouse duplicate genes are as essential as singletons. Trends Genet. 23, 378–381 (2007).
Segrè, D., Vitkup, D. & Church, G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA 99, 15112–15117 (2002).
Mo, M.L., Palsson, B.O. & Herrgard, M.J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 3, 37 (2009).
Kacser, H. & Burns, J.A. The molecular basis of dominance. Genetics 97, 639–666 (1981).
Brachmann, C.B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Amberg, D.C., Burke, D.J. & Strathern, J.N. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual 59–76 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2005).
Tong, A.H. et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 (2001).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Mumberg, D., Muller, R. & Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995).
Steinmetz, L.M. et al. Systematic screen for human disease genes in yeast. Nat. Genet. 31, 400–404 (2002).
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008).
Ekino, K., Kwon, I., Goto, M., Yoshino, S. & Furukawa, K. Functional analysis of HO gene in delayed homothallism in Saccharomyces cerevisiae wy2. Yeast 15, 451–458 (1999).
Meiron, H., Nahon, E. & Raveh, D. Identification of the heterothallic mutation in HO-endonuclease of S. cerevisiae using HO/ho chimeric genes. Curr. Genet. 28, 367–373 (1995).
Acknowledgements
We thank N. Bharucha, G. Kalay, A. Kumar, J. Ma and B. Williams for advice and assistance in yeast experiments, B. Palsson and his group for instruction on FBA, B.-Y. Liao for drawing Supplementary Figure 2 and M. Bakewell, S. Cho, W. Grus, B.-Y. Liao, and C. Maclean for valuable comments. This work was supported by research grants from the US National Institutes of Health and University of Michigan Center for Computational Medicine and Biology to J.Z.
Author information
Authors and Affiliations
Contributions
X.H. and J.Z. conceived the research; X.H., Z.W., W.Q. and J.Z. designed the experiments; X.H., W.Q., Z.W., Y.L. and J.Z. conducted the experiments; X.H., W.Q., Z.W. and J.Z. analyzed the data; X.H. and J.Z. drafted the manuscript and all authors contributed to the final manuscript writing and its revisions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–5 and Supplementary Note (PDF 3352 kb)
Rights and permissions
About this article
Cite this article
He, X., Qian, W., Wang, Z. et al. Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks. Nat Genet 42, 272–276 (2010). https://doi.org/10.1038/ng.524
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.524
This article is cited by
-
Transposon insertional mutagenesis of diverse yeast strains suggests coordinated gene essentiality polymorphisms
Nature Communications (2022)
-
Idiosyncratic epistasis creates universals in mutational effects and evolutionary trajectories
Nature Ecology & Evolution (2020)
-
Flux balance analysis with or without molecular crowding fails to predict two thirds of experimentally observed epistasis in yeast
Scientific Reports (2019)
-
Multiplexed deactivated CRISPR-Cas9 gene expression perturbations deter bacterial adaptation by inducing negative epistasis
Communications Biology (2018)
-
Evolutionary adaptations to new environments generally reverse plastic phenotypic changes
Nature Communications (2018)