Abstract
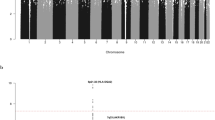
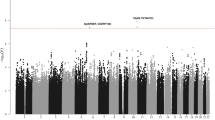
Using data from a genome-wide association study of 907 individuals with childhood acute lymphoblastic leukemia (cases) and 2,398 controls and with validation in samples totaling 2,386 cases and 2,419 controls, we have shown that common variation at 9p21.3 (rs3731217, intron 1 of CDKN2A) influences acute lymphoblastic leukemia risk (odds ratio = 0.71, P = 3.01 × 10−11), irrespective of cell lineage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Papaemmanuil, E. et al. Nat. Genet. 41, 1006–1010 (2009).
Treviño, L.R. et al. Nat. Genet. 41, 1001–1005 (2009).
Mullighan, C.G. & Downing, J.R. Leukemia 23, 1209–1218 (2009).
Bateman, C. et al. Blood published online. doi:10.1182/blood-2009-10-25143 (8 January 2010).
Falchi, M. et al. Nat. Genet. 41, 915–919 (2009).
Bishop, D.T. et al. Nat. Genet. 41, 920–925 (2009).
Stacey, S.N. et al. Nat. Genet. 41, 909–914 (2009).
Shete, S. et al. Nat. Genet. 41, 899–904 (2009).
Scott, L.J. et al. Science 316, 1341–1345 (2007).
McPherson, R. et al. Science 316, 1488–1491 (2007).
Sulong, S. et al. Blood 113, 100–107 (2009).
Mirebeau, D. et al. Haematologica 91, 881–885 (2006).
Stranger, B.E. et al. PLoS Genet. 1, e78 (2005).
Kitagawa, Y. et al. J. Biol. Chem. 277, 46289–46297 (2002).
Iacobucci, I. et al. Leukemia 24, 66–73 (2010).
Acknowledgements
The Kay Kendall Leukemia Fund and Leukemia Research (UK) provided principal funding for the study. Support from Cancer Research UK (C1298/A8362, supported by the Bobby Moore Fund) is also acknowledged. The study made use of genotyping data from the 1958 Birth Cohort which was generated and generously supplied to us by Panagiotis Deloukas of the Wellcome Trust Sanger Institute. For a full list of the investigators who contributed to the generation of the 1958 data, see URL section. We are grateful to S. Richards and J. Burrett (Clinical Trials Service Unit, Oxford) and L. Chilton (Leukaemia Research Cytogenetics Group, Northern Institute of Cancer Research, Newcastle Univ.), J. Simpson (Univ. York), P. Thomson and A. Hussain (Cancer Immunogenetics, School of Cancer Sciences, Univ. Manchester) for assistance with data harmonization. We thank the Leukaemia Research (LR) Childhood Cancer Leukaemia Group (CCLG) Childhood Leukaemia Cell Bank for access to the Medical Research Council ALL trial samples. We are grateful to the UK Cancer Cytogenetics Group (UKCCG) for data collection and provision of samples. P. Thomson (Cancer Immunogenetics, School of Cancer Sciences, University of Manchester) is funded by Children with Leukemia and we acknowledge their support. Genotyping of German cases and controls was partly covered by funding from Tumorzentrum Heidelberg-Mannheim, EU (Food-CT-2005-016320 and Health-F3-2007-2007-200767) and Deutsche Krebshilfe. The Hungarian sample collection was financially supported by grants from the Hungarian Scientific Research Fund (T042500), the Economic Competitiveness Operational Programme, Hungary (GVOP 3.1.1-2004-05-0022/3.0) and NKTH (National Research and Technology) TECH_08-A1/2-2008-0120. M.K. is a scholar of the Fonds de la Recherche en Santé du Québec. D.S. holds the François-Karl Viau Chair in Pediatric Oncogenomics and is also a scholar of the Fonds de la Recherche en Santé du Québec. J.H. is the recipient of a National Sciences and Engineering Research Council Canada Graduate's scholarship. P. Garcia-Miguel and A. Sastre from Hospital Infantil La Paz, Madrid, and J. Luis Vivanco Martínez from Hospital 12 de Octubre, Madrid, were involved in the sample collection of the Spanish samples, and R. Alonso from Centro Nacional de Investigaciones Oncológicas was involved in the genotyping.
We are grateful to all the study subjects and individuals for their participation and, finally, we would also like to thank the clinicians, other hospital staff and study staff who contributed to the blood sample and data collection for this study.
Author information
Authors and Affiliations
Contributions
R.S.H. and M.G. obtained financial support. R.S.H. designed and provided overall project management. R.S.H. drafted the manuscript with contributions from F.J.H., A.L.S. and M.G.; A.L.S. performed overall project management, development, database development and oversaw laboratory analyses; F.J.H. performed statistical analyses; F.J.H. and A.L.S. performed bioinformatics analyses; J.V. and E.P. performed UK sample preparation and genotyping; E.S. and S.E.K. performed curation and sample preparation of the Medical Research Council ALL 97 trial samples; T.L. and E.R. managed and maintained UKCCS sample data; M.T. performed curation and sample preparation of United Kingdom Childhood Cancer Study samples; J.M.A. and J.A.E.I. performed ascertainment, curation and sample preparation of the Northern Institute for Cancer Research case series. I.P.T. generated and managed UK colorectal cancer control genotypes. A.V.M. and C.J.H. performed UK CDKN2A deletion analysis; N.K., S.O. and H.P.K. carried out German CDKN2A deletion analysis; S.E.D. and Y.M. carried out HapMap and 1000 Genomes imputation; S.R. carried out survival analysis of UK data; M.Z. carried out survival analysis of German data; K.H. oversaw analysis of the German cohort; R.B.P., A.G. and R.K. conducted genotyping of German samples; R. Koehler, M. Stanulla, M. Schrappe and C.R.B. provided German DNA for analysis; D.J.E. and C.S. coordinated the data and sample collection of the Hungarian ALL cohort; A.F.S. genotyped the Hungarian samples; M.K., D.S. and J.H. performed curation and preparation of Canadian samples; A.G.N. was responsible for curation, management and genotyping of Spanish samples. All authors contributed to the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1–6 and Supplementary Figures 1–3. (PDF 1656 kb)
Rights and permissions
About this article
Cite this article
Sherborne, A., Hosking, F., Prasad, R. et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet 42, 492–494 (2010). https://doi.org/10.1038/ng.585
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.585
This article is cited by
-
Evaluation of rs10811661 polymorphism in CDKN2A / B in colon and gastric cancer
BMC Cancer (2023)
-
Association of CDKN2A/B mutations, PD-1, and PD-L1 with the risk of acute lymphoblastic leukemia in children
Journal of Cancer Research and Clinical Oncology (2023)
-
Association of IKZF1 and CDKN2A gene polymorphisms with childhood acute lymphoblastic leukemia: a high-resolution melting analysis
BMC Medical Genomics (2022)
-
Relationship between IKZF1 polymorphisms and the risk of acute lymphoblastic leukemia: a meta-analysis*
Oncology and Translational Medicine (2022)
-
Lymphoblastic Lymphoma: a Concise Review
Current Oncology Reports (2022)