Abstract
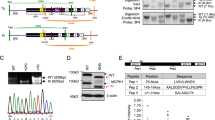
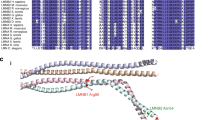
Genes associated with human microcephaly, a condition characterized by a small brain, include critical regulators of proliferation, cell fate and DNA repair. We describe a syndrome of congenital microcephaly and diverse defects in cerebral cortical architecture. Genome-wide linkage analysis in two families identified a 7.5-Mb locus on chromosome 19q13.12 containing 148 genes. Targeted high throughput sequence analysis of linked genes in each family yielded > 4,000 DNA variants and implicated a single gene, WDR62, as harboring potentially deleterious changes. We subsequently identified additional WDR62 mutations in four other families. Magnetic resonance imaging and postmortem brain analysis supports important roles for WDR62 in the proliferation and migration of neuronal precursors. WDR62 is a WD40 repeat–containing protein expressed in neuronal precursors as well as in postmitotic neurons in the developing brain and localizes to the spindle poles of dividing cells. The diverse phenotypes of WDR62 suggest it has central roles in many aspects of cerebral cortical development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sergi, C., Zoubaa, S. & Schiesser, M. Norman-Roberts syndrome: prenatal diagnosis and autopsy findings. Prenat. Diagn. 20, 505–509 (2000).
Albert, T.J. et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods 4, 903–905 (2007).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2009).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Desmet, F.-O. et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009).
Li, D. & Roberts, R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58, 2085–2097 (2001).
Hutchins, J.R.A. et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 (2010).
Bond, J. & Woods, C.G. Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 18, 95–101 (2006).
Rauch, A. et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319, 816–819 (2008).
Bond, J. et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37, 353–355 (2005).
Feng, Y. & Walsh, C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44, 279–293 (2004).
Lizarraga, S.B. et al. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137, 1907–1917 (2010).
Kouprina, N. et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum. Mol. Genet. 14, 2155–2165 (2005).
Thornton, G.K. & Woods, C.G. Primary microcephaly: do all roads lead to Rome? Trends Genet. 25, 501–510 (2009).
Wasserman, T. et al. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell 21, 117–130 (2010).
Saunders, R.D., Avides, M.C., Howard, T., Gonzalez, C. & Glover, D.M. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137, 881–890 (1997).
van der Voet, M. et al. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/G-α. Nat. Cell Biol. 11, 269–277 (2009).
Faulkner, N.E. et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2, 784–791 (2000).
Bond, J. et al. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32, 316–320 (2002).
Bilguvar, K. et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 (2010).
Hansen, D.V., Lui, J.H., Parker, P.R.L. & Kriegstein, A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010).
Gambello, M.J. et al. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J. Neurosci. 23, 1719–1729 (2003).
Sheen, V.L. et al. Impaired proliferation and migration in human Miller-Dieker neural precursors. Ann. Neurol. 60, 137–144 (2006).
Roberts, E. et al. Autosomal recessive primary microcephaly: an analysis of locus heterogeneity and phenotypic variation. J. Med. Genet. 39, 718–721 (2002).
Gul, A. et al. Genetic studies of autosomal recessive primary microcephaly in 33 Pakistani families: novel sequence variants in ASPM gene. Neurogenetics 7, 105–110 (2006).
Roberts, E. et al. The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1–13.2. Eur. J. Hum. Genet. 7, 815–820 (1999).
den Dunnen, J.T. & Antonarakis, S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 15, 7–12 (2000).
Major, J.E. Genomic mutation consequence calculator. Bioinformatics 23, 3091–3092 (2007).
Kent, W.J. et al. The Human Genome Browser at UCSC. Genome Res. 12, 996–1006 (2002).
Tucker, E.S. et al. Molecular specification and patterning of progenitor cells in the lateral and medial ganglionic eminences. J. Neurosci. 28, 9504–9518 (2008).
Acknowledgements
We thank the many families and researchers who participated in this study; B. Chang and A. Poduri for expert review of MRI imaging findings; S. Lizarraga for contributions to genetic mapping; M. Aita and the In situ Hybridization Core Facility, University of North Carolina Neuroscience Center, Chapel Hill, North Carolina for performing in situ hybridizations; C.G. Woods for helpful discussions and for sharing data before publication; A. von Deimling from the Institut für Pathologie, Heidelberg, for sectioning of the LIS-2601 brain; A. Nicholas and M. Khurshid for advice regarding immunohistochemistry; A. Hill and L. Bu at the MRDDRC (Mental Retardation Developmental Disability Research Center) Imaging Core for assistance with confocal microscopy (US National Institutes of Health (NIH) grant P30-HD-18655 and Fidelity Foundation); and R. Sean Hill for assistance with linkage analysis and helpful discussions. We are indebted to the members of the Microcephaly Collaborative (Supplementary Table 1) for contributing to the cohort from which these families were drawn. T.W.Y. was supported by a NIH T32 grant (T32 NS007484-08), the Clinical Investigator Training Program (CITP) at Harvard-Massachusetts Institute of Technology Health Sciences and Technology and Beth Israel Deaconess Medical Center in collaboration with Pfizer, Inc. and Merck and Company, Inc., and the Nancy Lurie Marks Junior Faculty MeRIT Fellowship. G.H.M. was supported by the Young Investigator Award of National Alliance for Research on Schizophrenia and Depression (NARSAD) as a NARSAD Lieber Investigator. Research was supported by grants from the National Institute of Neurological Disorders and Stroke (NINDS) (RO1 NSR01-35129) and the Fogarty International Center (R21 NS061772) to C.A.W., the Dubai Harvard Foundation for Medical Research and the Manton Center for Orphan Disease Research. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the NIH to Johns Hopkins University, contract number HHSN268200782096C and NIH N01-HG-65403. Genotyping at Children's Hospital Boston was supported by the Intellectual and Developmental Disabilities Research Centers (CHB DDRC, P30 HD18655). Genotyping at the Broad Institute was supported by National Human Genome Research Institute (NHGRI). C.A.W. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
T.W.Y. helped characterize MCSG syndrome, designed and performed targeted capture experiments, generated sequencing libraries, designed bioinformatics pipelines, analyzed sequencing data, identified WDR62 mutations, designed WDR62 in situ expression studies, designed, performed and analyzed WDR62 immunohistochemistry studies, helped analyze the LIS-2601 postmortem specimen and wrote the manuscript. G.H.M. helped characterize MCSG syndrome, performed initial genome wide linkage studies and identified the MCSG locus. D.J.T. designed and performed WDR62 immunohistochemistry studies, helped analyze in situ expression studies and helped analyze the LIS-2601 postmortem specimen. S.K.S. helped characterize MCSG syndrome, identified additional affected families, analyzed SNP and microsatellite genotyping and narrowed the MCSG genomic interval. L.F.-S. helped characterize MCSG syndrome and wrote the initial clinical description of the LIS-900 family. C.M.S. identified the LIS-2600 family and provided clinical information and the LIS-2601 postmortem specimen. M.T. identified the MC-1400 and MC-1600 families and provided clinical information. M.T.M. identified the PH-16900 family and provided clinical information. B.J.B. organized clinical information and subject samples. J.M.F. sequenced WDR62 in families to confirm high throughput sequencing findings and to discover additional alleles. C.S. helped with bioinformatic pipelines and generated constructs for WDR62 in situ expression studies. W.B.D. referred the PH-16900 family. R.D.F. analyzed the LIS-2601 postmortem specimen. A.J.B. reviewed MRIs for characterization of MCSG syndrome. C.A.W. directed the overall research, helped analyze the LIS-2601 postmortem specimen and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–4 and Supplementary Table 1 (PDF 2835 kb)
Supplementary Movie 1
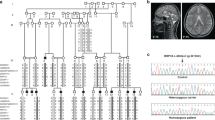
Microcephaly, simplified gyri, polymicrogyria, and schizencephaly in a patient with a WDR62 mutation. T1-weighted MRI sequence of a patient (PH-16903) with a homozygous V65M mutation, demonstrating microcephaly, simplified gyri, relatively preserved brain stem and cerebellum, widespread polymicrogyria, and right temporo-parietal open lip schizencephaly. (MOV 464 kb)
Rights and permissions
About this article
Cite this article
Yu, T., Mochida, G., Tischfield, D. et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet 42, 1015–1020 (2010). https://doi.org/10.1038/ng.683
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.683
This article is cited by
-
WDR62-deficiency Causes Autism-like Behaviors Independent of Microcephaly in Mice
Neuroscience Bulletin (2023)
-
Novel phenotype and genotype spectrum of WDR62 in two patients with associated primary autosomal recessive microcephaly
Irish Journal of Medical Science (1971 -) (2022)
-
WDR62 is required for centriole duplication in spermatogenesis and manchette removal in spermiogenesis
Communications Biology (2021)
-
Primary microcephaly with an unstable genome
Genome Instability & Disease (2020)
-
Deconstructing cortical folding: genetic, cellular and mechanical determinants
Nature Reviews Neuroscience (2019)