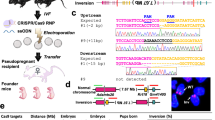
Abstract
Embryonic stem cell technology revolutionized biology by providing a means to assess mammalian gene function in vivo. Although it is now routine to generate mice from embryonic stem cells, one of the principal methods used to create mutations, gene targeting, is a cumbersome process. Here we describe the indexing of 93,960 ready-made insertional targeting vectors from two libraries. 5,925 of these vectors can be used directly to inactivate genes with an average targeting efficiency of 28%. Combinations of vectors from the two libraries can be used to disrupt both alleles of a gene or engineer larger genomic changes such as deletions, duplications, translocations or inversions. These indexed vectors constitute a public resource (Mutagenic Insertion and Chromosome Engineering Resource; MICER) for high-throughput, targeted manipulation of the mouse genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Waterston, R.H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Collins, F.S., Green, E.D. & Guyer, M.S. A vision for the future of genomics research. Nature 422, 835–847 (2003).
Evans, M.J. & Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981).
Bradley, A., Evans, M., Kaufman, M.H. & Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256 (1984).
Thomas, K.R. & Capecchi, M.R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503–512 (1987).
Smithies, O., Gregg, R.G., Boggs, S.S., Koralewski, M.A. & Kucherlapati, R.S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317, 230–234 (1985).
Bult, C.J. et al. The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res. 32, D476–D481 (2004).
Skarnes, W.C. et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 36, 543–544 (2004).
Gossler, A., Joyner, A.L., Rossant, J. & Skarnes, W.C. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science 244, 463–465 (1989).
Hasty, P., Crist, M., Grompe, M. & Bradley, A. Efficiency of insertion versus replacement vector targeting varies at different chromosomal loci. Mol. Cell Biol. 14, 8385–8390 (1994).
Zheng, B., Mills, A.A. & Bradley, A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 11, 2354–2360 (1999).
Mills, A.A. et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 32, 708–713 (1999).
te Riele, H., Maandag, E.R., Clarke, A., Hooper, M. & Berns, A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature 348, 649–651 (1990).
Yu, Y. & Bradley, A. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2, 780–790 (2001).
Ramirez-Solis, R., Liu, P. & Bradley, A. Chromosome engineering in mice. Nature 378, 720–724 (1995).
Smith, A.J. et al. A site-directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination. Nat. Genet. 9, 376–385 (1995).
Birney, E. et al. Ensembl 2004. Nucleic Acids Res. 32, D468–D470 (2004).
Testa, G. et al. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 21, 443–447 (2003).
Liu, P., Jenkins, N.A. & Copeland, N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 (2003).
Kucera, G.T., Bortner, D.M. & Rosenberg, M.P. Overexpression of an Agouti cDNA in the skin of transgenic mice recapitulates dominant coat color phenotypes of spontaneous mutants. Dev Biol. 173, 162–173 (1996).
Yokoyama, T. et al. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 18, 7293–7298 (1990).
Zheng, B., Vogel, H., Donehower, L.A. & Bradley, A. Visual genotyping of a coat color tagged p53 mutant mouse line. Cancer Biol. Ther. 1, 433–435 (2002).
Nishijima, I., Mills, A., Qi, Y., Mills, M. & Bradley, A. Two new balancer chromosomes on mouse chromosome 4 to facilitate functional annotation of human chromosome 1p. Genesis 36, 142–148 (2003).
Walz, K. et al. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol. Cell. Biol. 23, 3646–3655 (2003).
Klysik, J., Dinh, C. & Bradley, A. Two new mouse chromosome 11 balancers. Genomics 83, 303–310 (2004).
Ning, Z., Cox, A.J. & Mullikin, J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001).
Acknowledgements
We thank W.C. Skarnes, members of the laboratories of W.C. Skarnes and A.B. and the Sanger informatics team for discussions and assistance. D.J.A and L.v.d.W. were supported by a National Health and Medical Research Council CJ Martin fellowship and a National Health and Medical Research Council CJ Martin/RG Menizes fellowship, respectively. J.J. is a recipient of a NWO Genomics Fellowship from the Netherlands Organization for Scientific Research. This work was supported by the Wellcome Trust (UK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Targeting frequency data for MICER vectors. (XLS 13 kb)
Supplementary Table 2
Sentinel genes list. (XLS 1040 kb)
Supplementary Table 3
MICER gene disruption alleles. (XLS 1328 kb)
Supplementary Table 4
Restriction enzymes that can be used to linearize MICER vectors. (PDF 3 kb)
Rights and permissions
About this article
Cite this article
Adams, D., Biggs, P., Cox, T. et al. Mutagenic Insertion and Chromosome Engineering Resource (MICER). Nat Genet 36, 867–871 (2004). https://doi.org/10.1038/ng1388
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1388
This article is cited by
-
Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE
Scientific Reports (2017)
-
Deletions linked to TP53 loss drive cancer through p53-independent mechanisms
Nature (2016)
-
Chd5 orchestrates chromatin remodelling during sperm development
Nature Communications (2014)
-
Engineered chromosome-based genetic mapping establishes a 3.7 Mb critical genomic region for Down syndrome-associated heart defects in mice
Human Genetics (2014)