Abstract
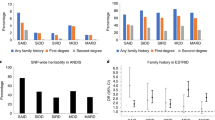
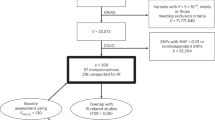
Genome-wide association studies are now identifying disease-associated chromosome regions. However, even after convincing replication, the localization of the causal variant(s) requires comprehensive resequencing, extensive genotyping and statistical analyses in large sample sets leading to targeted functional studies. Here, we have localized the type 1 diabetes (T1D) association in the interleukin 2 receptor alpha (IL2RA) gene region to two independent groups of SNPs, spanning overlapping regions of 14 and 40 kb, encompassing IL2RA intron 1 and the 5′ regions of IL2RA and RBM17 (odds ratio = 2.04, 95% confidence interval = 1.70–2.45; P = 1.92 × 10−28; control frequency = 0.635). Furthermore, we have associated IL2RA T1D susceptibility genotypes with lower circulating levels of the biomarker, soluble IL-2RA (P = 6.28 × 10−28), suggesting that an inherited lower immune responsiveness predisposes to T1D.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wang, W.Y., Barratt, B.J., Clayton, D.G. & Todd, J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 6, 109–118 (2005).
Dewan, A. et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 314, 989–992 (2006).
Duerr, R.H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Todd, J.A. Statistical false positive or true disease pathway? Nat. Genet. 38, 731–733 (2006).
Todd, J.A. et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 (2007).
Frayling, T.M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Ueda, H. et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423, 506–511 (2003).
Vella, A. et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 76, 773–779 (2005).
Qu, H.Q., Montpetit, A., Ge, B., Hudson, T.J. & Polychronakos, C. Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes 56, 1174–1176 (2007).
Brand, O.J. et al. Association of the interleukin-2 receptor alpha (IL-2Ralpha)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin. Endocrinol. 66, 508–512 (2007).
Fehniger, T.A. & Caligiuri, M.A. Interleukin 15: biology and relevance to human disease. Blood 97, 14–32 (2001).
Waldmann, T.A. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 6, 595–601 (2006).
Kuniyasu, Y. et al. Naturally anergic and suppressive CD25(+)CD4(+) T cells as a functionally and phenotypically distinct immunoregulatory T cell subpopulation. Int. Immunol. 12, 1145–1155 (2000).
Yamanouchi, J. et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat. Genet. 39, 329–337 (2007).
Robb, R.J. & Kutny, R.M. Structure-function relationships for the IL 2-receptor system. IV. Analysis of the sequence and ligand-binding properties of soluble Tac protein. J. Immunol. 139, 855–862 (1987).
Bleesing, J. et al. The diagnostic significance of soluble CD163 and soluble interleukin-2 receptor alpha-chain in macrophage activation syndrome and untreated new-onset systemic juvenile idiopathic arthritis. Arthritis Rheum. 56, 965–971 (2007).
Makis, A.C. et al. Serum levels of soluble interleukin-2 receptor alpha (sIL-2Ralpha) as a predictor of outcome in brucellosis. J. Infect. 51, 206–210 (2005).
Kim, H.P., Imbert, J. & Leonard, W.J. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 17, 349–366 (2006).
The International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
The International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005).
Clayton, D.G. et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 37, 1243–1246 (2005).
Hardenbol, P. et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21, 673–678 (2003).
Spielman, R.S., McGinnis, R.E. & Ewens, W.J. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 52, 506–516 (1993).
Klose, R.J. & Bird, A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006).
Roh, T.Y., Cuddapah, S., Cui, K. & Zhao, K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 103, 15782–15787 (2006).
Bell, G.I., Horita, S. & Karam, J.H. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 33, 176–183 (1984).
Naranjo-Gomez, M. et al. Expression and function of the IL-2 receptor in activated human plasmacytoid dendritic cells. Eur. J. Immunol 37, 1764–1772 (2007).
Giordano, C. et al. Interleukin 2 and soluble interleukin 2-receptor secretion defect in vitro in newly diagnosed type I diabetic patients. Diabetes 38, 310–315 (1989).
Leonard, W.J. et al. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature 311, 626–631 (1984).
Tree, T.I., Roep, B.O. & Peakman, M. A mini meta-analysis of studies on CD4+CD25+ T cells in human type 1 diabetes: report of the Immunology of Diabetes Society T Cell Workshop. Ann. NY Acad. Sci. 1079, 9–18 (2006).
Rubinstein, M.P. et al. Converting IL-15 to a superagonist by binding to soluble IL-15RA. Proc. Natl. Acad. Sci. USA 103, 9166–9171 (2006).
Power, C. & Elliott, J. Cohort profile: 1958 British birth cohort (National Child Development Study). Int. J. Epidemiol. 35, 34–41 (2006).
Bain, S.C., Todd, J.A. & Barnett, A.H. The British Diabetic Association–Warren repository. Autoimmunity 7, 83–85 (1990).
Feltbower, R.G., McKinney, P.A., Parslow, R.C., Stephenson, C.R. & Bodansky, H.J. Type 1 diabetes in Yorkshire, UK: time trends in 0–14 and 15–29-year-olds, age at onset and age-period-cohort modelling. Diabet. Med. 20, 437–441 (2003).
Lernmark, A. et al. Family cell lines available for research. Am. J. Hum. Genet. 47, 1028–1030 (1990).
Patterson, C.C., Carson, D.J. & Hadden, D.R. Epidemiology of childhood IDDM in Northern Ireland 1989–1994: low incidence in areas with highest population density and most household crowding. Northern Ireland Diabetes Study Group. Diabetologia 39, 1063–1069 (1996).
Smyth, D.J. et al. Assessing the validity of the association between the SUMO4 M55V variant and risk of type 1 diabetes. Nat. Genet. 37, 110–111 (2005).
Undlien, D.E. et al. No difference in the parental origin of susceptibility HLA class II haplotypes among Norwegian patients with insulin-dependent diabetes mellitus. Am. J. Hum. Genet. 57, 1511–1514 (1995).
Ionescu-Tirgoviste, C. et al. Low frequency of HLA DRB1*03 - DQB1*02 and DQB1*0302 haplotypes in Romania is consistent with the country's low incidence of Type I diabetes. Diabetologia 44 (Suppl. 3), B60–B66 (2001).
Smink, L.J. et al. T1DBase, a community web-based resource for type 1 diabetes research. Nucleic Acids Res. 33, D544–D549 (2005).
Bonfield, J.K., Rada, C. & Staden, R. Automated detection of point mutations using fluorescent sequence trace subtraction. Nucleic Acids Res. 26, 3404–3409 (1998).
She, J.X. et al. Heterophile antibodies segregate in families and are associated with protection from type 1 diabetes. Proc. Natl. Acad. Sci. USA 96, 8116–8119 (1999).
Plagnol, V., Cooper, J.D., Todd, J.A. & Clayton, D.G. A method to address differential bias in genotyping in large-scale association studies. PLoS Genet. [online] 3, e74 (2007)(doi:10.1371/journal.pgen.0030074).
Chapman, J.M., Cooper, J.D., Todd, J.A. & Clayton, D.G. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum. Hered. 56, 18–31 (2003).
Scheet, P. & Stephens, M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644 (2006).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Burren, O.S. et al. Development of an integrated genome informatics, data management and workflow infrastructure: a toolbox for the study of complex disease genetics. Hum. Genomics 1, 98–109 (2004).
Cordell, H.J. & Clayton, D.G. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am. J. Hum. Genet. 70, 124–141 (2002).
Acknowledgements
We acknowledge the participation of all the affected individuals, control subjects and family members and thank the Human Biological Data Interchange and Diabetes UK for the US and UK multiplex families, respectively; the Norwegian Study Group for Childhood Diabetes for the collection of Norwegian families (D. Undlien and K. Rønningen); C. Ionescu-Tîrgovişte and C. Guja for the Romanian samples; T. Valle, J. Tuomilehto and V. Harjutsalo for the Finnish samples and D. Savage, C. Patterson and P. Maxwell for the Northern Irish samples. We acknowledge use of the DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council Grant G0000934 and Wellcome Trust Grant 068545/Z/02 and thank D. Strachan and P. Burton for their help. We also thank The Avon Longitudinal Study of Parents and Children laboratory in Bristol, including S. Ring and W. McArdle for preparing and providing the control DNA samples. We thank T. Willis, M. Faham and P. Hardenbol of Affymetrix for the molecular inversion probe technology. We thank L. Smink, O. Burren, B. Healy, V. Everett and G. Dolman for bioinformatics and IT support. DNA samples were prepared by T. Mistry, T. Hassanali, M. Maisuria, M. Sebastian, S. Wood, P. Lauder and M. Hardy together with G. Coleman, E. King, A. Simpson and H. Stevens, who also prepared the plasma samples with the assistance of H. Schuilenburg. This work was funded by the Wellcome Trust and the Juvenile Diabetes Research Foundation (JDRF) International, the American Diabetes Association (grant 7-06-RA-09) awarded to M.A., as well as funding from the JDRF International Pilot and Feasibility Grant awarded to T.B. and M.A. (7-2006-328).
Author information
Authors and Affiliations
Contributions
C.E.L. contributed to the conception, design and coordination of the study, regional annotation, SNP discovery, genotyping, sIL-2RA studies, data analysis and drafting of the manuscript. J.D.C. contributed to the study design, data analysis and drafting of the manuscript. N.M.W. managed the data. D.J.S. and R.B. contributed to SNP discovery and genotyping, K.B. contributed to the plasma sIL-2RA study. S.F. contributed to the genotyping. V.P. and D.G.C. contributed to data analysis. T.B., L.S.W. and M.A. contributed to the design, coordination and analysis of the sIL-2RA studies. J.A.T. participated in the conception, design and coordination of the study, data analysis and drafting of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5, Supplementary Figure 1 (PDF 165 kb)
Rights and permissions
About this article
Cite this article
Lowe, C., Cooper, J., Brusko, T. et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet 39, 1074–1082 (2007). https://doi.org/10.1038/ng2102
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng2102
This article is cited by
-
Unsolved mystery of Fas: mononuclear cells may have trouble dying in patients with Sjögren’s syndrome
BMC Immunology (2023)
-
Whole exome sequencing identified a novel splice donor site variant in interleukin 2 receptor alpha chain
Immunogenetics (2023)
-
The link between circulating follicular helper T cells and autoimmunity
Nature Reviews Immunology (2022)
-
The heterogeneous pathogenesis of type 1 diabetes mellitus
Nature Reviews Endocrinology (2019)
-
Pre-diagnostic genotyping identifies T1D subjects with impaired Treg IL-2 signaling and an elevated proportion of FOXP3+IL-17+ cells
Genes & Immunity (2017)