Abstract
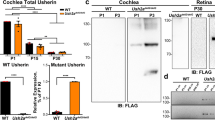
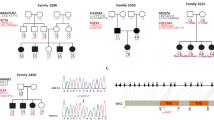
Hearing impairment affects about 1 in 1,000 children at birth. Approximately 70 loci implicated in non-syndromic forms of deafness have been reported in humans and 24 causative genes have been identified1 (see also http://www.uia.ac.be/dnalab/hhh). We report a mouse transcript, isolated by a candidate deafness gene approach, that is expressed almost exclusively in the inner ear. Genomic analysis shows that the human ortholog STRC (so called owing to the name we have given its protein—stereocilin), which is located on chromosome 15q15, contains 29 exons encompassing approximately 19 kb. STRC is tandemly duplicated, with the coding sequence of the second copy interrupted by a stop codon in exon 20. We have identified two frameshift mutations and a large deletion in the copy containing 29 coding exons in two families affected by autosomal recessive non-syndromal sensorineural deafness linked to the DFNB16 locus. Stereocilin is made up of 1,809 amino acids, and contains a putative signal petide and several hydrophobic segments. Using immunohistolabeling, we demonstrate that, in the mouse inner ear, stereocilin is expressed only in the sensory hair cells and is associated with the stereocilia, the stiff microvilli forming the structure for mechanoreception of sound stimulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Petit, C., Levilliers, J. & Hardelin, J.P. Molecular genetics of hearing loss. Annu. Rev. Genet. (in press).
Hubank, M. & Schatz, D.G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 22, 5640–5648 (1994).
Cohen-Salmon, M., El-Amraoui, A., Leibovici, M. & Petit, C. Otogelin: a glycoprotein specific to the acellular membranes of the inner ear. Proc. Natl Acad. Sci. USA 94, 14450–14455 (1997).
Verpy, E., Leibovici, M. & Petit, C. Characterization of otoconin-95, the major protein of murine otoconia, provides insights into the formation of these inner ear biominerals. Proc. Natl Acad. Sci. USA 96, 529–534 (1999).
Verpy, E. et al. A defect in harmonin, a PDZ domain–containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nature Genet. 26, 51–55 (2000).
Yasunaga, S. et al. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nature Genet. 21, 363–369 (1999).
Simmler, M.C. et al. Targeted disruption of Otog results in deafness and severe imbalance. Nature Genet. 24, 139–143 (2000).
Simmler, M.C. et al. Twister mutant mice are defective for otogelin, a component specific to inner ear acellular membranes. Mamm. Genome 11, 961–966 (2000).
Campbell, D.A. et al. A new locus for non-syndromal, autosomal recessive, sensorineural hearing loss (DFNB16) maps to human chromosome 15q21-q22. J. Med. Genet. 34, 1015–1017 (1997).
Villamar, M., del Castillo, I., Valle, N., Romero, L. & Moreno, F. Deafness locus DFNB16 is located on chromosome 15q13–q21 within a 5-cM interval flanked by markers D15S994 and D15S132. Am. J. Hum. Genet. 64, 1238–1241 (1999).
Kozak, M. Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome 7, 563–574 (1996).
von Heijne, G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14, 4683–4690 (1986).
Popot, J.L. & de Vitry, C. On the microassembly of integral membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 19, 369–403 (1990).
Zine, A. & Romand, R. Development of the auditory receptors of the rat: a SEM study. Brain Res. 721, 49–58 (1996).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159 (1987).
Klein, P., Kanehisa, M. & DeLisi, C. The detection and classification of membrane-spanning proteins. Biochim. Biophys. Acta 815, 468–476 (1985).
Küssel-Andermann, P. et al. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J. 19, 6020–6029 (2000).
el-Amraoui, A. et al. Human Usher 1B/mouse shaker-1: the retinal phenotype discrepancy explained by the presence/absence of myosin VIIA in the photoreceptor cells. Hum. Mol. Genet. 5, 1171–1178 (1996).
Engelman, D.M., Steitz, T.A. & Goldman, A. Identifying nonpolar transbilayer helices in amino-acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15, 321–353 (1986).
Acknowledgements
We thank S. Chardenoux for aid with drafting figures. This work was supported by grants from the European Economic Community (QLG2-CT-1999-00988), Fondation pour la Recherche Médicale, National Lotteries Charity Board through Defeating Deafness, the Royal National Institute for Deaf People, the Wellcome Trust, and fundings from the CAICYT (SAF99-0025) of Spanish Ministerio de la Ciencia and FIS 00-0244 of Spanish Ministerio de Sanidad. I.Z. is a recipient from Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Verpy, E., Masmoudi, S., Zwaenepoel, I. et al. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet 29, 345–349 (2001). https://doi.org/10.1038/ng726
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng726
This article is cited by
-
MPZL2—a common autosomal recessive deafness gene related to moderate sensorineural hearing loss in the Chinese population
BMC Medical Genomics (2024)
-
The genetic basis and the diagnostic yield of genetic testing related to nonsyndromic hearing loss in Qatar
Scientific Reports (2024)
-
Genetic etiology of non-syndromic hearing loss in Europe
Human Genetics (2022)
-
ARNSHL gene identification: past, present and future
Molecular Genetics and Genomics (2022)
-
Identification of autosomal recessive nonsyndromic hearing impairment genes through the study of consanguineous and non-consanguineous families: past, present, and future
Human Genetics (2022)