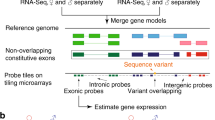
Abstract
Here we present a statistically rigorous approach to quantifying microarray expression data that allows the relative effects of multiple classes of treatment to be compared and incorporates analytical methods that are common to quantitative genetics. From the magnitude of gene effects and contributions of variance components, we find that gene expression in adult flies is affected most strongly by sex, less so by genotype and only weakly by age (for 1- and 6-wk flies); in addition, sex × genotype interactions may be present for as much as 10% of the Drosophila transcriptome. This interpretation is compromised to some extent by statistical issues relating to power and experimental design. Nevertheless, we show that changes in expression as small as 1.2-fold can be highly significant. Genotypic contributions to transcriptional variance may be of a similar magnitude to those relating to some quantitative phenotypes and should be considered when assessing the significance of experimental treatments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lynch, M. & Walsh, B. Genetics and Analysis of Quantitative Traits (Sinauer Associates, Sunderland, MA, 1998).
Eisen, M.B. & Brown, P.O. DNA arrays for analysis of gene expression. Methods Enzymol. 303, 179–205 (1999).
Wolfinger, R.D. et al. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. (in press).
Kerr, M.K. & Churchill, G.A. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77, 123–128 (2001).
Kerr, M.K., Martin, M. & Churchill, G.A. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7, 819–837 (2001).
Sandberg, R. et al. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc. Natl Acad. Sci. USA 97, 11038–11043 (2000).
Primig, M. et al. The core meiotic transcriptome in budding yeasts. Nature Genet. 26, 415–423 (2000).
Spicer, G.S. Genetic differentiation of Drosophila melanogaster populations as assessed by two-dimensional electrophoresis. Biochem. Genet. 29, 389–401 (1991).
Nuzhdin, S., Pasyukova, E.G., Dilda, C., Zeng, Z.-B. & Mackay, T.F.C. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 94, 9734–9739 (1997).
Chapman, T. & Partridge, L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with males. Proc. R. Soc. Lond. B 263, 755–759.
Webster, G.C. & Webster, S.L. Specific disappearance of translatable messenger RNA for elongation factor one in aging Drosophila melanogaster. Mech. Ageing. Dev. 24, 335–342 (1984).
Zou, S., Meadows, S., Sharp, L., Jan, L.Y. & Jan, Y.N. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 97, 13726–13731 (2000).
Leips, J. & Mackay, T.F.C. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155, 1773–1788 (2000).
Littell, R.C., Milliken, G.A., Stroup, W.W. & Wolfinger, R.D. SAS System for Mixed Models (SAS Institute, Cary, NC, 1996).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998).
Cochran, W.G. & Cox, G.M. Experimental Designs (Wiley, New York, 1957).
SAS Institute Inc. SAS/STAT Software Version 8 (SAS Institute, Cary, NC, 1999).
Hughes, T.R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Perrimon, N., Lanjuin, A., Arnold, C. & Noll, E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics 144, 1681–1692 (1996).
Andrews, J. et al. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10, 2030–2043 (2000).
White, K.P., Rifkin, S.A., Hurban, P. & Hogness, D.S. Microarray analysis of Drosophila development during metamorphosis. Science 286, 2179–2184 (1999).
Long, A.D., Lyman, R.F., Morgan, A.H., Langley, C.H. & Mackay, T.F.C. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154,1255–1269 (2000).
Ross, D.T. et al. Systematic variation in gene expression patterns in human cancer cell lines. Nature Genetics 24, 227–235 (2000).
Hiller, M.A., Lin, T.Y., Wood, C. & Fuller, M.T. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15, 1021–1030 (2001).
Mace, M.L. Jr, Montagu, J., Rose, S.D. & McGuinness, G. in Microarray Biochip Technology (ed. Schena, M.) 39–64 (Eaton Publishers, Natick, MA, 2000).
Acknowledgements
We thank J. Sall for discussions; B. Sosinski and L. He for help with resequencing the EST set and establishing the microarray facility; and M. Arbeitman, B. Null, E. Johnson, E. Furlong, F. Imam, A. Wagoner and M. Magwire for helping to prepare miniprep cDNA clones from the EST library. This work was supported by grants to G.G. from the National Institute of Aging and the David and Lucille Packard Foundation.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Jin, W., Riley, R., Wolfinger, R. et al. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet 29, 389–395 (2001). https://doi.org/10.1038/ng766
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng766
This article is cited by
-
Detection Algorithms for Simple Two-Group Comparisons Using Spontaneous Reporting Systems
Drug Safety (2024)
-
De novo transcriptome assembly of Dalbergia sissoo Roxb. (Fabaceae) under Botryodiplodia theobromae-induced dieback disease
Scientific Reports (2023)
-
Identification of Genes Involved in Digestion from Transcriptome of Parasesarma pictum and Parasesarma affine Hepatopancreas
Thalassas: An International Journal of Marine Sciences (2022)
-
Species diversity and spatial distribution of CL/VL vectors: assessing bioclimatic effect on expression plasticity of genes possessing vaccine properties isolated from wild-collected sand flies in endemic areas of Iran
BMC Infectious Diseases (2021)
-
Transcriptome profiling of Lymnaea stagnalis (Gastropoda) for ecoimmunological research
BMC Genomics (2021)