Abstract
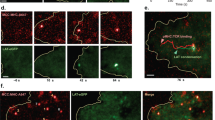
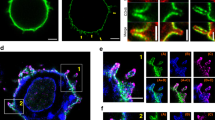
The organization and dynamics of receptors and other molecules in the plasma membrane are not well understood. Here we analyzed the spatio-temporal dynamics of T cell antigen receptor (TCR) complexes and linker for activation of T cells (Lat), a key adaptor molecule in the TCR signaling pathway, in T cell membranes using high-speed photoactivated localization microscopy, dual-color fluorescence cross-correlation spectroscopy and transmission electron microscopy. In quiescent T cells, both molecules existed in separate membrane domains (protein islands), and these domains concatenated after T cell activation. These concatemers were identical to signaling microclusters, a prominent hallmark of T cell activation. This separation versus physical juxtapositioning of receptor domains and domains containing downstream signaling molecules in quiescent versus activated T cells may be a general feature of plasma membrane–associated signal transduction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
29 January 2010
In the version of this article initially published, some rows in Table 1 were misaligned. The error has been corrected in the HTML and PDF versions of the article.
References
Monks, C.R., Freiberg, B.A., Kupfer, H., Sciaky, N. & Kupfer, A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395, 82–86 (1998).
Grakoui, A. et al. The immunological synapse: a molecular machine controlling T cell activation. Science 285, 221–227 (1999).
Bunnell, S.C. et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 158, 1263–1275 (2002).
Campi, G., Varma, R. & Dustin, M.L. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202, 1031–1036 (2005).
Yokosuka, T. et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 (2005).
Varma, R., Campi, G., Yokosuka, T., Saito, T. & Dustin, M.L. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25, 117–127 (2006).
Wilson, B.S., Pfeiffer, J.R. & Oliver, J.M. Observing FcεRI signaling from the inside of the mast cell membrane. J. Cell Biol. 149, 1131–1142 (2000).
Wilson, B.S., Pfeiffer, J.R., Surviladze, Z., Gaudet, E.A. & Oliver, J.M. High resolution mapping of mast cell membranes reveals primary and secondary domains of FcεRI and LAT. J. Cell Biol. 154, 645–658 (2001).
Schamel, W.W. et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 202, 493–503 (2005).
Fernandez-Miguel, G. et al. Multivalent structure of an αβT cell receptor. Proc. Natl. Acad. Sci. USA 96, 1547–1552 (1999).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S.T. et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA 104, 17370–17375 (2007).
Shroff, H., Galbraith, C.G., Galbraith, J.A. & Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 (2008).
Magde, D., Elson, E.L. & Webb, W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers 13, 29–61 (1974).
Bacia, K., Kim, S.A. & Schwille, P. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods 3, 83–89 (2006).
Groves, J.T., Parthasarathy, R. & Forstner, M.B. Fluorescence imaging of membrane dynamics. Annu. Rev. Biomed. Eng. 10, 311–338 (2008).
Sanan, D.A. & Anderson, R.G. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J. Histochem. Cytochem. 39, 1017–1024 (1991).
Lillemeier, B.F., Pfeiffer, J.R., Surviladze, Z., Wilson, B.S. & Davis, M.M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. USA 103, 18992–18997 (2006).
Chudakov, D.M. et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat. Biotechnol. 22, 1435–1439 (2004).
Seder, R.A., Paul, W.E., Davis, M.M. & Fazekas de St Groth, B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 176, 1091–1098 (1992).
Ripley, B.D. Modeling spatial patterns. J. R. Stat. Soc. [Ser A] B39, 172–212 (1977).
Ripley, B.D. Tests of randomness for spatial point patterns. J. R. Stat. Soc. [Ser A] B41, 368–374 (1979).
Liu, J. et al. Crystallization of a deglycosylated T cell receptor (TCR) complexed with an anti-TCR Fab fragment. J. Biol. Chem. 271, 33639–33646 (1996).
Fahmy, T.M., Bieler, J.G., Edidin, M. & Schneck, J.P. Increased TCR avidity after T cell activation: a mechanism for sensing low-density antigen. Immunity 14, 135–143 (2001).
Aivazian, D. & Stern, L.J. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat. Struct. Biol. 7, 1023–1026 (2000).
Sigalov, A.B., Aivazian, D.A., Uversky, V.N. & Stern, L.J. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry 45, 15731–15739 (2006).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008).
Sako, Y. & Kusumi, A. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125, 1251–1264 (1994).
Kusumi, A., Ike, H., Nakada, C., Murase, K. & Fujiwara, T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 17, 3–21 (2005).
James, J.R. et al. Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc. Natl. Acad. Sci. USA 104, 17662–17667 (2007).
Simson, R., Sheets, E.D. & Jacobson, K. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys. J. 69, 989–993 (1995).
Irvine, D.J., Purbhoo, M.A., Krogsgaard, M. & Davis, M.M. Direct observation of ligand recognition by T cells. Nature 419, 845–849 (2002).
Exley, M., Wileman, T., Mueller, B. & Terhorst, C. Evidence for multivalent structure of T cell antigen receptor complex. Mol. Immunol. 32, 829–839 (1995).
Bonefeld, C.M. et al. TCR comodulation of nonengaged TCR takes place by a protein kinase C and CD3γ di-leucine-based motif-dependent mechanism. J. Immunol. 171, 3003–3009 (2003).
Douglass, A.D. & Vale, R.D. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell 121, 937–950 (2005).
Chu, D.H., Morita, C.T. & Weiss, A. The Syk family of protein tyrosine kinases in T cell activation and development. Immunol. Rev. 165, 167–180 (1998).
Samelson, L.E. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol. 20, 371–394 (2002).
Zhang, J. et al. Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron 37, 14–34 (2006).
Forstner, M.B., Yee, C.K., Parikh, A.N. & Groves, J.T. Lipid lateral mobility and membrane phase structure modulation by protein binding. J. Am. Chem. Soc. 128, 15221–15227 (2006).
Acknowledgements
We thank B. Wilson for advice on TEM and plasma membrane sheet preparation; J. Perrino from the Cell Sciences Imaging Facility for expertise and service; and F. Tynan for comments on the manuscript. Supported by the US National Institutes of Health (AI 55277 to M.M.D.), the US National Science Foundation (J.T.G.), the Howard Hughes Medical Institute (M.M.D. and J.T.G.) and the Human Frontier Science Program (B.F.L.).
Author information
Authors and Affiliations
Contributions
B.F.L. and M.M.D., conceptualization and manuscript preparation; B.F.L., PALM experiments and analysis, TEM studies, molecular biology, cell biology and protein chemistry; M.A.M., PALM conceptualization and analysis software; M.B.F. and J.T.G., dcFCCS conceptualization; M.B.F., dcFCCS software and analysis; M.B.F. and B.F.L., dcFCCS experiments; and J.B.H., anti-TCRβ-scFv and microscope setup.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–2 and Supplementary Methods (PDF 8659 kb)
Rights and permissions
About this article
Cite this article
Lillemeier, B., Mörtelmaier, M., Forstner, M. et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol 11, 90–96 (2010). https://doi.org/10.1038/ni.1832
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1832
This article is cited by
-
LFA-1 nanoclusters integrate TCR stimulation strength to tune T-cell cytotoxic activity
Nature Communications (2024)
-
Spectroscopic single-molecule localization microscopy: applications and prospective
Nano Convergence (2023)
-
Nanoparticle-based modulation of CD4+ T cell effector and helper functions enhances adoptive immunotherapy
Nature Communications (2022)
-
Approach to map nanotopography of cell surface receptors
Communications Biology (2022)
-
Correction of multiple-blinking artifacts in photoactivated localization microscopy
Nature Methods (2022)