Abstract
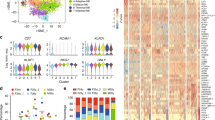
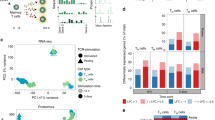
Using whole-genome microarray data sets of the Immunological Genome Project, we demonstrate a closer transcriptional relationship between NK cells and T cells than between any other leukocytes, distinguished by their shared expression of genes encoding molecules with similar signaling functions. Whereas resting NK cells are known to share expression of a few genes with cytotoxic CD8+ T cells, our transcriptome-wide analysis demonstrates that the commonalities extend to hundreds of genes, many encoding molecules with unknown functions. Resting NK cells demonstrate a 'preprimed' state compared with naive T cells, which allows NK cells to respond more rapidly to viral infection. Collectively, our data provide a global context for known and previously unknown molecular aspects of NK cell identity and function by delineating the genome-wide repertoire of gene expression of NK cells in various states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Heng, T.S. & Painter, M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Bendelac, A., Bonneville, M. & Kearney, J.F. Autoreactivity by design: innate B and T lymphocytes. Nat. Rev. Immunol. 1, 177–186 (2001).
Vivier, E. et al. Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49 (2011).
Herberman, R.B., Nunn, M.E., Holden, H.T. & Lavrin, D.H. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int. J. Cancer 16, 230–239 (1975).
Kiessling, R., Klein, E., Pross, H. & Wigzell, H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur. J. Immunol. 5, 117–121 (1975).
Lanier, L.L. & Phillips, J.H. Ontogeny of natural killer cells. Nature 319, 269–270 (1986).
Di Santo, J.P. Natural killer cell developmental pathways: a question of balance. Annu. Rev. Immunol. 24, 257–286 (2006).
Yamagata, T., Benoist, C. & Mathis, D. A shared gene-expression signature in innate-like lymphocytes. Immunol. Rev. 210, 52–66 (2006).
Chambers, S.M. et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell 1, 578–591 (2007).
Obata-Onai, A. et al. Comprehensive gene expression analysis of human NK cells and CD8+ T lymphocytes. Int. Immunol. 14, 1085–1098 (2002).
Hanna, J. et al. Novel insights on human NK cells' immunological modalities revealed by gene expression profiling. J. Immunol. 173, 6547–6563 (2004).
Dybkaer, K. et al. Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics 8, 230 (2007).
Lanier, L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9, 495–502 (2008).
Bezman, N. & Koretzky, G.A. Compartmentalization of ITAM and integrin signaling by adapter molecules. Immunol. Rev. 218, 9–28 (2007).
Pont, F. et al. The gene expression profile of phosphoantigen-specific human gammadelta T lymphocytes is a blend of αβ T-cell and NK-cell signatures. Eur. J. Immunol. 42, 228–240 (2012).
Wilson, S.B. & Byrne, M.C. Gene expression in NKT cells: defining a functionally distinct CD1d-restricted T cell subset. Curr. Opin. Immunol. 13, 555–561 (2001).
Hesslein, D.G. & Lanier, L.L. Transcriptional control of natural killer cell development and function. Adv. Immunol. 109, 45–85 (2011).
Sun, Y. et al. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 34, 6314–6326 (2006).
Li, M.O. & Flavell, R.A. TGF-β: a master of all T cell trades. Cell 134, 392–404 (2008).
Akbulut, S. et al. Sprouty proteins inhibit receptor-mediated activation of phosphatidylinositol-specific phospholipase C. Mol. Biol. Cell 21, 3487–3496 (2010).
Narni-Mancinelli, E. et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA 108, 18324–18329 (2011).
Yu, J. et al. NKp46 identifies an NKT cell subset susceptible to leukemic transformation in mouse and human. J. Clin. Invest. 121, 1456–1470 (2011).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Walzer, T. et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat. Immunol. 8, 1337–1344 (2007).
Despoix, N. et al. Mouse CD146/MCAM is a marker of natural killer cell maturation. Eur. J. Immunol. 38, 2855–2864 (2008).
Arase, H., Saito, T., Phillips, J.H. & Lanier, L.L. Cutting edge: the mouse NK cell-associated antigen recognized by DX5 monoclonal antibody is CD49b (α2 integrin, very late antigen-2). J. Immunol. 167, 1141–1144 (2001).
Ewen, C.L., Kane, K.P. & Bleackley, R.C. A quarter century of granzymes. Cell Death Differ. 19, 28–35 (2012).
Young, J.D., Hengartner, H., Podack, E.R. & Cohn, Z.A. Purification and characterization of a cytolytic pore-forming protein from granules of cloned lymphocytes with natural killer activity. Cell 44, 849–859 (1986).
Colige, A. et al. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J. Biol. Chem. 277, 5756–5766 (2002).
Hirst, C.E. et al. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 170, 805–815 (2003).
Zhang, M. et al. Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity 24, 451–461 (2006).
Liu, L. et al. A novel protein tyrosine kinase NOK that shares homology with platelet-derived growth factor/fibroblast growth factor receptors induces tumorigenesis and metastasis in nude mice. Cancer Res. 64, 3491–3499 (2004).
Kaech, S.M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002).
Kallies, A., Xin, A., Belz, G.T. & Nutt, S.L. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31, 283–295 (2009).
Shin, H. et al. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009).
Sun, J.C., Lopez-Verges, S., Kim, C.C., DeRisi, J.L. & Lanier, L.L. NK cells and immune “memory”. J. Immunol 186, 1891–1897 (2011).
Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146 (2003).
Palmer, D.C. & Restifo, N.P. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 30, 592–602 (2009).
Lee, S.H., Kim, K.S., Fodil-Cornu, N., Vidal, S.M. & Biron, C.A. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J. Exp. Med. 206, 2235–2251 (2009).
Sun, J.C., Beilke, J.N. & Lanier, L.L. Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009).
Xue, L., Chiang, L., He, B., Zhao, Y.Y. & Winoto, A. FoxM1, a forkhead transcription factor is a master cell cycle regulator for mouse mature T cells but not double positive thymocytes. PLoS ONE 5, e9229 (2011).
Zhou, M. et al. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J. Immunol. 178, 5496–5504 (2007).
O'Leary, J.G., Goodarzi, M., Drayton, D.L. & von Andrian, U.H. T cell- and B cell- independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 7, 507–516 (2006).
Sun, J.C. & Lanier, L.L. NK cell development, homeostasis and function: parallels with CD8 T cells. Nat. Rev. Immunol. 11, 645–(2011).
Cui, W. & Kaech, S.M. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol. Rev. 236, 151–166 (2010).
Wherry, E.J. et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007).
Albrecht, I. et al. Persistence of effector memory Th1 cells is regulated by Hopx. Eur. J. Immunol. 40, 2993–3006 (2010).
Hawiger, D., Wan, Y.Y., Eynon, E.E. & Flavell, R.A. The transcription cofactor Hopx is required for regulatory T cell function in dendritic cell-mediated peripheral T cell unresponsiveness. Nat. Immunol. 11, 962–968 (2010).
Malhotra, D. et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat. Immunol. 13, 499–510 (2012).
Acknowledgements
We thank A. Weiss (University of California, San Francisco) for antibody to Syk; the members of the ImmGen Consortium and M. Dozmorov for discussions; the ImmGen core team (M. Painter, J. Ericson and S. Davis) for data generation and processing; J. Jarjoura and J. Arakawa-Hoyt for assistance in cell sorting; A. Beaulieu, J. Karo and S. Madera for data from MCMV infection experiments; and eBioscience, Affymetrix and Expression Analysis for support of the ImmGen Project. Supported by the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (R24 AI072073 and R01 AI068129; T32AI060537 to D.W.H.; T32AI060536 to J.A.B.; and AI072117 to A.W.G.), the American Cancer Society (L.L.L. and N.A.B.), the Canadian Institutes of Health Research (G.M.-O.) and the Searle Scholars Program (J.C.S.).
Author information
Authors and Affiliations
Consortia
Contributions
C.C.K. analyzed data; N.A.B. and J.C.S. sorted cell subsets, did follow-up experiments and analyzed data; G.M.-O., D.W.H. and Y.K. did experiments; J.A.B. and A.W.G. designed and did the T cell studies; and N.A.B., C.C.K., J.C.S. and L.L.L. designed studies and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Full list of members and affiliations appears at the end of the paper.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5 and Notes 1–2 (PDF 3341 kb)
Rights and permissions
About this article
Cite this article
Bezman, N., Kim, C., Sun, J. et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol 13, 1000–1009 (2012). https://doi.org/10.1038/ni.2395
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2395
This article is cited by
-
A diversity of novel type-2 innate lymphoid cell subpopulations revealed during tumour expansion
Communications Biology (2024)
-
The transcription factor Fli1 restricts the formation of memory precursor NK cells during viral infection
Nature Immunology (2022)
-
Sequential actions of EOMES and T-BET promote stepwise maturation of natural killer cells
Nature Communications (2021)
-
Innate lymphocytes: pathogenesis and therapeutic targets of liver diseases and cancer
Cellular & Molecular Immunology (2021)
-
PD-1 blockade-unresponsive human tumor-infiltrating CD8+ T cells are marked by loss of CD28 expression and rescued by IL-15
Cellular & Molecular Immunology (2021)