Abstract
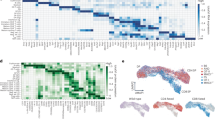
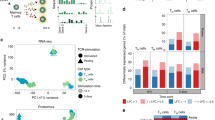
The differentiation of αβT cells from thymic precursors is a complex process essential for adaptive immunity. Here we exploited the breadth of expression data sets from the Immunological Genome Project to analyze how the differentiation of thymic precursors gives rise to mature T cell transcriptomes. We found that early T cell commitment was driven by unexpectedly gradual changes. In contrast, transit through the CD4+CD8+ stage involved a global shutdown of housekeeping genes that is rare among cells of the immune system and correlated tightly with expression of the transcription factor c-Myc. Selection driven by major histocompatibility complex (MHC) molecules promoted a large-scale transcriptional reactivation. We identified distinct signatures that marked cells destined for positive selection versus apoptotic deletion. Differences in the expression of unexpectedly few genes accompanied commitment to the CD4+ or CD8+ lineage, a similarity that carried through to peripheral T cells and their activation, demonstrated by mass cytometry phosphoproteomics. The transcripts newly identified as encoding candidate mediators of key transitions help define the 'known unknowns' of thymocyte differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
Change history
13 May 2013
In the version of this article initially published online, the eighth and ninth author names were incorrect. Those should be Matthew H. Spitzer and Garry P. Nolan. The error has been corrected for the print, PDF and HTML versions of this article.
References
Miller, J.F. The golden anniversary of the thymus. Nat. Rev. Immunol. 11, 489–495 (2011).
von Boehmer, H., Teh, H.S. & Kisielow, P. The thymus selects the useful, neglects the useless and destroys the harmful. Immunol. Today 10, 57–61 (1989).
Jameson, S.C., Hogquist, K.A. & Bevan, M.J. Positive selection of thymocytes. Annu. Rev. Immunol. 13, 93–126 (1995).
Robey, E. Regulation of T cell fate by Notch. Annu. Rev. Immunol. 17, 283–295 (1999).
Hogquist, K.A., Baldwin, T.A. & Jameson, S.C. Central tolerance: learning self-control in the thymus. Nat. Rev. Immunol. 5, 772–782 (2005).
Stritesky, G.L., Jameson, S.C. & Hogquist, K.A. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol. 30, 95–114 (2012).
Singer, A., Adoro, S. & Park, J.H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788–801 (2008).
Thompson, P.K. & Zuniga-Pflucker, J.C. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin. Immunol. 23, 350–359 (2011).
Rothenberg, E.V. T cell lineage commitment: identity and renunciation. J. Immunol. 186, 6649–6655 (2011).
Godfrey, D.I., Kennedy, J., Suda, T. & Zlotnik, A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150, 4244–4252 (1993).
Bhandoola, A., von, B.H., Petrie, H.T. & Zuniga-Pflucker, J.C. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 26, 678–689 (2007).
Williams, J.A. et al. Regulated costimulation in the thymus is critical for T cell development: dysregulated CD28 costimulation can bypass the pre-TCR checkpoint. J. Immunol. 175, 4199–4207 (2005).
Prinz, I. et al. Visualization of the earliest steps of γδ T cell development in the adult thymus. Nat. Immunol. 7, 995–1003 (2006).
Kreslavsky, T. et al. β-Selection-induced proliferation is required for αβ T cell differentiation. Immunity 37, 840–853 (2012).
Zinkernagel, R.M., Callahan, G.N., Klein, J. & Dennert, G. Cytotoxic T cells learn specificity for self H-2 during diffentiation in the thymus. Nature 271, 251–253 (1978).
MacDonald, H.R. et al. Positive selection of CD4+ thymocytes controlled by MHC class II gene products. Nature 336, 471–473 (1988).
Kisielow, P., Teh, H.S., Bluthmann, H. & von Boehmer, H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature 335, 730–733 (1988).
Bousso, P., Bhakta, N.R., Lewis, R.S. & Robey, E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science 296, 1876–1880 (2002).
Egerton, M., Scollay, R. & Shortman, K. Kinetics of mature T-cell development in the thymus. Proc. Natl. Acad. Sci. USA 87, 2579–2582 (1990).
Teh, H.S. et al. Thymic major histocompatibility complex antigens and the αβ T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature 335, 229–233 (1988).
Jordan, M.S. et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2, 301–306 (2001).
Leishman, A.J. et al. Precursors of functional MHC class I- or class II-restricted CD8αα+ T cells are positively selected in the thymus by agonist self-peptides. Immunity 16, 355–364 (2002).
Fink, P.J. & Hendricks, D.W. Post-thymic maturation: young T cells assert their individuality. Nat. Rev. Immunol. 11, 544–549 (2011).
DeRyckere, D., Mann, D.L. & DeGregori, J. Characterization of transcriptional regulation during negative selection in vivo. J. Immunol. 171, 802–811 (2003).
Schmitz, I., Clayton, L.K. & Reinherz, E.L. Gene expression analysis of thymocyte selection in vivo. Int. Immunol. 15, 1237–1248 (2003).
Liston, A. et al. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity 21, 817–830 (2004).
Mick, V.E. et al. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J. Immunol. 173, 5434–5444 (2004).
Zucchelli, S. et al. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity 22, 385–396 (2005).
Baldwin, T.A. & Hogquist, K.A. Transcriptional analysis of clonal deletion in vivo. J. Immunol. 179, 837–844 (2007).
Mingueneau, M. et al. Thymic negative selection is functional in NOD mice. J. Exp. Med. 209, 623–637 (2012).
Zhang, J.A. et al. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467–482 (2012).
Li, L., Leid, M. & Rothenberg, E.V. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93 (2010).
Luc, S. et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat. Immunol. 13, 412–419 (2012).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Narayan, K. et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat. Immunol. 13, 511–518 (2012).
Ikawa, T. et al. An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 (2010).
Li, P. et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 (2010).
Rothenberg, E.V., Zhang, J. & Li, L. Multilayered specification of the T-cell lineage fate. Immunol. Rev. 238, 150–168 (2010).
Rakhilin, S.V. et al. A network of control mediated by regulator of calcium/calmodulin-dependent signaling. Science 306, 698–701 (2004).
Kisielow, J., Nairn, A.C. & Karjalainen, K. TARPP, a novel protein that accompanies TCR gene rearrangement and thymocyte education. Eur. J. Immunol. 31, 1141–1149 (2001).
Olenchock, B.A. et al. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy. Nat. Immunol. 7, 1174–1181 (2006).
Guo, R. et al. Synergistic control of T cell development and tumor suppression by diacylglycerol kinase α and ζ. Proc. Natl. Acad. Sci. USA 105, 11909–11914 (2008).
Deftos, M.L. et al. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13, 73–84 (2000).
Reizis, B. & Leder, P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 16, 295–300 (2002).
González-Garcia, S. et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7Rα gene expression in early human thymopoiesis and leukemia. J. Exp. Med. 206, 779–791 (2009).
Yashiro-Ohtani, Y. et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 23, 1665–1676 (2009).
Taghon, T. et al. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006).
Germar, K. et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl. Acad. Sci. USA 108, 20060–20065 (2011).
Weber, B.N. et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011).
Weng, A.P. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109 (2006).
von Boehmer, H. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv. Immunol. 84, 201–238 (2004).
Wong, G.W. et al. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRβ-selected mouse thymocytes. Blood 120, 1439–1448 (2012).
Dose, M. et al. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood 108, 2669–2677 (2006).
Choi, S.H., Wright, J.B., Gerber, S.A. & Cole, M.D. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 24, 1236–1241 (2010).
Adhikary, S. & Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6, 635–645 (2005).
Schreiber-Agus, N. & DePinho, R.A. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays 20, 808–818 (1998).
Ciofani, M. et al. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J. Immunol. 172, 5230–5239 (2004).
Maillard, I. et al. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 203, 2239–2245 (2006).
Iritani, B.M. et al. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 21, 4820–4830 (2002).
Geering, B. & Simon, H.U. Peculiarities of cell death mechanisms in neutrophils. Cell Death Differ. 18, 1457–1469 (2011).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Lovén, J. et al. Revisiting global gene expression analysis. Cell 151, 476–482 (2012).
Neilson, J.R., Zheng, G.X., Burge, C.B. & Sharp, P.A. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21, 578–589 (2007).
Starr, T.K., Jameson, S.C. & Hogquist, K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003).
Swat, W., Dessing, M., von, B.H. & Kisielow, P. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur. J. Immunol. 23, 739–746 (1993).
Jones, M.E. & Zhuang, Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27, 860–870 (2007).
Rivera, R.R. et al. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity 12, 17–26 (2000).
Bain, G. et al. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat. Immunol. 2, 165–171 (2001).
Meissner, T.B. et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. USA 107, 13794–13799 (2010).
Huang, B., Wu, P., Popov, K.M. & Harris, R.A. Starvation and diabetes reduce the amount of pyruvate dehydrogenase phosphatase in rat heart and kidney. Diabetes 52, 1371–1376 (2003).
Maliekal, P. et al. Molecular identification of mammalian phosphopentomutase and glucose-1,6-bisphosphate synthase, two members of the α-D-phosphohexomutase family. J. Biol. Chem. 282, 31844–31851 (2007).
van Meerwijk, J.P. et al. Quantitative impact of thymic clonal deletion on the T cell repertoire. J. Exp. Med. 185, 377–383 (1997).
Surh, C.D. & Sprent, J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 372, 100–103 (1994).
Viret, C. et al. A role for accessibility to self-peptide-self-MHC complexes in intrathymic negative selection. J. Immunol. 166, 4429–4437 (2001).
McCaughtry, T.M., Baldwin, T.A., Wilken, M.S. & Hogquist, K.A. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J. Exp. Med. 205, 2575–2584 (2008).
Baldwin, K.K., Trenchak, B.P., Altman, J.D. & Davis, M.M. Negative selection of T cells occurs throughout thymic development. J. Immunol. 163, 689–698 (1999).
Krueger, A. & von Boehmer, H. Identification of a T lineage-committed progenitor in adult blood. Immunity 26, 105–116 (2007).
Liston, A. et al. Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: genomic analysis by mRNA profiling. Genome Biol. 8, R12 (2007).
Chan, C.J., Andrews, D.M. & Smyth, M.J. Receptors that interact with nectin and nectin-like proteins in the immunosurveillance and immunotherapy of cancer. Curr. Opin. Immunol. 24, 246–251 (2012).
Wang, L. & Bosselut, R. CD4–CD8 lineage differentiation: Thpok-ing into the nucleus. J. Immunol. 183, 2903–2910 (2009).
Pénit, C. & Vasseur, F. Expansion of mature thymocyte subsets before emigration to the periphery. J. Immunol. 159, 4848–4856 (1997).
Lei, Y. & Takahama, Y. XCL1 and XCR1 in the immune system. Microbes Infect. 14, 262–267 (2012).
Rudolph, B., Hueber, A.O. & Evan, G.I. Reversible activation of c-Myc in thymocytes enhances positive selection and induces proliferation and apoptosis in vitro. Oncogene 19, 1891–1900 (2000).
Yu, S. et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826 (2012).
Tullai, J.W. et al. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J. Biol. Chem. 282, 23981–23995 (2007).
Bouillet, P. et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 (1999).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Eden, E., Lipson, D., Yogev, S. & Yakhini, Z. Discovering motifs in ranked lists of DNA sequences. PLOS Comput. Biol. 3, e39 (2007).
Eden, E. et al. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Kanehisa, M. et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114 (2012).
Acknowledgements
We thank K. Hattori, A. Ortiz-Lopez and N. Asinovski for technical assistance with mice and antibodies; J. Moore and A. Kressler for flow cytometry; and C. Laplace for assistance with figure preparation. Supported by the US National Institutes of Health (AI072073), the Human Frontier Science Program (HFSP-LT000096 to M.M.), the National Health and Medical Research Council (637353 to D.G.) and the Australian Research Council (LP110201169 to, T.H.).
Author information
Authors and Affiliations
Consortia
Contributions
M.M., T.K., D.G. and T.H. designed, did and analyzed some of the experiments and wrote the manuscript; R.C. and J.E. helped with microarray data analysis; S.B. and M.H.S. designed, did and analyzed some of the experiments; G.P.N. and K.K. and H.v.B. contributed resources and assisted with data analysis; and D.M. and C.B. directed the study, analyzed and interpreted results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations appears at the end of the paper.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1987 kb)
Supplementary Table 1
Supplementary Table 1 (XLSX 10 kb)
Supplementary Table 2
Supplementary Table 2 (XLSX 596 kb)
Supplementary Table 3
Supplementary Table 3 (XLSX 262 kb)
Supplementary Table 4
Supplementary Table 4 (XLSX 355 kb)
Rights and permissions
About this article
Cite this article
Mingueneau, M., Kreslavsky, T., Gray, D. et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol 14, 619–632 (2013). https://doi.org/10.1038/ni.2590
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2590
This article is cited by
-
Expression of E-cadherin by CD8+ T cells promotes their invasion into biliary epithelial cells
Nature Communications (2024)
-
Mannosylated glycans impair normal T-cell development by reprogramming commitment and repertoire diversity
Cellular & Molecular Immunology (2023)
-
MNT suppresses T cell apoptosis via BIM and is critical for T lymphomagenesis
Cell Death & Differentiation (2023)
-
The β-selection step shapes T-cell identity
Nature (2023)
-
Pre-T cell receptor self-MHC sampling restricts thymocyte dedifferentiation
Nature (2023)