Abstract
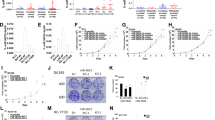
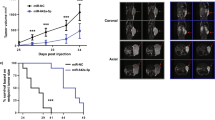
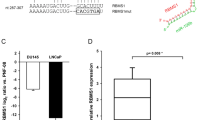
MicroRNAs (miRNAs) are noncoding small RNAs that repress protein translation by targeting specific messenger RNAs. miR-15a and miR-16-1 act as putative tumor suppressors by targeting the oncogene BCL2. These miRNAs form a cluster at the chromosomal region 13q14, which is frequently deleted in cancer. Here, we report that the miR-15a and miR-16-1 cluster targets CCND1 (encoding cyclin D1) and WNT3A, which promotes several tumorigenic features such as survival, proliferation and invasion. In cancer cells of advanced prostate tumors, the miR-15a and miR-16 level is significantly decreased, whereas the expression of BCL2, CCND1 and WNT3A is inversely upregulated. Delivery of antagomirs specific for miR-15a and miR-16 to normal mouse prostate results in marked hyperplasia, and knockdown of miR-15a and miR-16 promotes survival, proliferation and invasiveness of untransformed prostate cells, which become tumorigenic in immunodeficient NOD-SCID mice. Conversely, reconstitution of miR-15a and miR-16-1 expression results in growth arrest, apoptosis and marked regression of prostate tumor xenografts. Altogether, we propose that miR-15a and miR-16 act as tumor suppressor genes in prostate cancer through the control of cell survival, proliferation and invasion. These findings have therapeutic implications and may be exploited for future treatment of prostate cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jemal, A. et al. Cancer statistics, 2006. CA Cancer J. Clin. 56, 106–130 (2006).
Loberg, R.D., Logothetis, C.J., Keller, E.T. & Pienta, K.J. Pathogenesis and treatment of prostate cancer bone metastases: targeting the lethal phenotype. J. Clin. Oncol. 23, 8232–8241 (2005).
Pienta, K.J. & Smith, D.C. Advances in prostate cancer chemotherapy: a new era begins. CA Cancer J. Clin. 55, 300–318 (2005).
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Care, A. et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618 (2007).
Calin, G.A. et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353, 1793–1801 (2005).
Calin, G.A. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 101, 2999–3004 (2004).
Esquela-Kerscher, A. & Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 (2006).
Calin, G.A. et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99, 15524–15529 (2002).
Dong, J.T., Boyd, J.C. & Frierson, H.F. Jr. Loss of heterozygosity at 13q14 and 13q21 in high-grade, high-stage prostate cancer. Prostate 49, 166–171 (2001).
Hyytinen, E.R., Frierson, H.F., Jr., Boyd, J.C., Chung, L.W. & Dong, J.T. Three distinct regions of allelic loss at 13q14, 13q21–22, and 13q33 in prostate cancer. Genes Chromosom. Cancer 25, 108–114 (1999).
Cimmino, A. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 102, 13944–13949 (2005).
Yin, Z. et al. Limiting the location of a putative human prostate cancer tumor suppressor gene at chromosome 13q14.3. Oncogene 18, 7576–7583 (1999).
Ebert, M.S., Neilson, J.R. & Sharp, P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 (2007).
Nupponen, N.N., Hyytinen, E.R., Kallioniemi, A.H. & Visakorpi, T. Genetic alterations in prostate cancer cell lines detected by comparative genomic hybridization. Cancer Genet. Cytogenet. 101, 53–57 (1998).
Bonci, D. et al. 'Advanced' generation lentiviruses as efficient vectors for cardiomyocyte gene transduction in vitro and in vivo. Gene Ther. 10, 630–636 (2003).
Felli, N. et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. USA 102, 18081–18086 (2005).
Follenzi, A., Ailles, L.E., Bakovic, S., Geuna, M. & Naldini, L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25, 217–222 (2000).
Clevers, H. Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 (2006).
Dhanasekaran, S.M. et al. Delineation of prognostic biomarkers in prostate cancer. Nature 412, 822–826 (2001).
Sherr, C.J. Cancer cell cycles. Science 274, 1672–1677 (1996).
Almeida, M., Han, L., Bellido, T., Manolagas, S.C. & Kousteni, S. Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by β-catenin–dependent and –independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J. Biol. Chem. 280, 41342–41351 (2005).
Yun, M.S., Kim, S.E., Jeon, S.H., Lee, J.S. & Choi, K.Y. Both ERK and Wnt/β-catenin pathways are involved in WNT3A-induced proliferation. J. Cell Sci. 118, 313–322 (2005).
Bello, D., Webber, M.M., Kleinman, H.K., Wartinger, D.D. & Rhim, J.S. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis 18, 1215–1223 (1997).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 438, 685–689 (2005).
Tolcher, A.W. et al. A phase II, pharmacokinetic, and biological correlative study of oblimersen sodium and docetaxel in patients with hormone-refractory prostate cancer. Clin. Cancer Res. 11, 3854–3861 (2005).
Zimmermann, T.S. et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006).
Navone, N.M., Olive, M. & Troncoso, P. Isolation and culture of prostate cancer cell lines. Methods Mol. Med. 88, 121–132 (2004).
Nelson, P.T. et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 12, 187–191 (2006).
Lewis, B.P., Burge, C.B. & Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P. & Burge, C.B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003).
John, B. et al. Human MicroRNA targets. PLoS Biol. 2, e363 (2004).
Krek, A. et al. Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 (2005).
Acknowledgements
We thank G. Loreto, A. Di Virgilio and M. Blasi for technical assistance and E. Palio and M. Gulisano for valuable discussion and suggestions. This work was supported by the Italian Health Ministry, the Italian Ministry of University and Research (project number RBIP06ZJ78), and the Italian Association for Cancer Research. M. Bartucci is a recipient of a fellowship from the Italian Association for Cancer Research.
Author information
Authors and Affiliations
Contributions
D.B., V.C., M.M., A.A., R.G., L.M., L.D., A.P., M. Biffoni, C.L. and M. Bartucci conducted the in vitro and in vivo experiments. G.M., D.B. and R.D.M. planned the experiments. L.D. provided human samples. G.M. and C.P. discussed the results and commented on the manuscript. R.D.M. wrote the manuscript and was responsible for research coordination and strategy.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–11, Supplementary Tables 1 and 2 and Supplementary Data and Methods (PDF 1753 kb)
Rights and permissions
About this article
Cite this article
Bonci, D., Coppola, V., Musumeci, M. et al. The miR-15a–miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14, 1271–1277 (2008). https://doi.org/10.1038/nm.1880
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.1880
This article is cited by
-
Reprogramming of palmitic acid induced by dephosphorylation of ACOX1 promotes β-catenin palmitoylation to drive colorectal cancer progression
Cell Discovery (2023)
-
Learning noncoding RNA biology from viruses
Mammalian Genome (2022)
-
Immunomodulation by Bifidobacterium animalis subsp. lactis Bb12: Integrative Analysis of miRNA Expression and TLR2 Pathway–Related Target Proteins in Swine Monocytes
Probiotics and Antimicrobial Proteins (2022)
-
Epigenetic effects toward new insights as potential therapeutic target in B-thalassemia
Journal of Genetic Engineering and Biotechnology (2021)
-
The roles of microRNAs in mouse development
Nature Reviews Genetics (2021)