Abstract
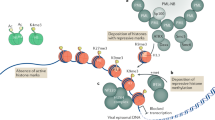
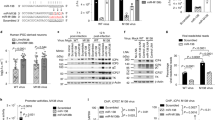
Reversible methylation of histone tails serves as either a positive signal recognized by transcriptional assemblies or a negative signal that result in repression1,2,3,4. Invading viral pathogens that depend upon the host cell's transcriptional apparatus are also subject to the regulatory impact of chromatin assembly and modifications5,6,7,8. Here we show that infection by the α-herpesviruses, herpes simplex virus (HSV) and varicella zoster virus (VZV), results in the rapid accumulation of chromatin bearing repressive histone H3 Lys9 methylation. To enable expression of viral immediate early (IE) genes, both viruses use the cellular transcriptional coactivator host cell factor-1 (HCF-1) to recruit the lysine-specific demethylase-1 (LSD1) to the viral immediate early promoters. Depletion of LSD1 or inhibition of its activity with monoamine oxidase inhibitors (MAOIs) results in the accumulation of repressive chromatin and a block to viral gene expression. As HCF-1 is a component of the Set1 and MLL1 histone H3 Lys4 methyltransferase complexes9,10, it thus coordinates modulation of repressive H3 Lys9 methylation levels with addition of activating H3 Lys4 trimethylation marks. Strikingly, MAOIs also block the reactivation of HSV from latency in sensory neurons, indicating that the HCF-1 complex is a crucial component of the reactivation mechanism. The results support pharmaceutical control of histone modifying enzymes as a strategy for controlling herpesvirus infections.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Klose, R.J. & Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318 (2007).
Ruthenburg, A.J., Allis, C.D. & Wysocka, J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 (2007).
Shi, Y. & Whetstine, J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 25, 1–14 (2007).
Kouzarides, T. & Berger, S.L. Chromatin modifications and their mechanism of action. in Epigenetics (eds. Allis, C.D., Jenuwein, T. & Reinberg, D.) 191–209 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2007).
Knipe, D.M. & Cliffe, A. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6, 211–221 (2008).
Lieberman, P.M. Chromatin organization and virus gene expression. J. Cell. Physiol. 216, 295–302 (2008).
Mok, H.P. & Lever, A.M. Chromatin, gene silencing and HIV latency. Genome Biol. 8, 228 (2007).
Sinclair, J. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta published online, doi:10.1016/j.bbagrm.2009.08.001 (12 August 2009).
Wysocka, J., Myers, M.P., Laherty, C.D., Eisenman, R.N. & Herr, W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3–K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17, 896–911 (2003).
Yokoyama, A. et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24, 5639–5649 (2004).
Kristie, T.M. Early events pre-initiation of alphaherpesvirus viral gene expression. in Human Herpesviruses Biology, Therapy, and Immunoprophylaxis (eds. Arvin, A. et al.) 112–127 (Cambridge University Press, New York, 2007).
Narayanan, A., Ruyechan, W.T. & Kristie, T.M. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. USA 104, 10835–10840 (2007).
Huang, J. et al. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 80, 5740–5746 (2006).
Garcia-Bassets, I. et al. Histone methylation–dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128, 505–518 (2007).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Perillo, B. et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319, 202–206 (2008).
Wang, J. et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887 (2007).
Dou, Y. et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 (2006).
Lee, M.G., Wynder, C., Cooch, N. & Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005).
Shi, Y.J. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005).
Forneris, F., Binda, C., Vanoni, M.A., Mattevi, A. & Battaglioli, E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 579, 2203–2207 (2005).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).
Lee, M.G., Wynder, C., Schmidt, D.M., McCafferty, D.G. & Shiekhattar, R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13, 563–567 (2006).
Schmidt, D.M. & McCafferty, D.G. trans-2-phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46, 4408–4416 (2007).
Yang, M. et al. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry 46, 8058–8065 (2007).
Shin, S. & Janknecht, R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 359, 742–746 (2007).
Wissmann, M. et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor–dependent gene expression. Nat. Cell Biol. 9, 347–353 (2007).
Amelio, A.L., Giordani, N.V., Kubat, N.J., O'Neil, J.E. & Bloom, D.C. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80, 2063–2068 (2006).
Kubat, N.J., Tran, R.K., McAnany, P. & Bloom, D.C. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78, 1139–1149 (2004).
Neumann, D.M., Bhattacharjee, P.S., Giordani, N.V., Bloom, D.C. & Hill, J.M. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 81, 13248–13253 (2007).
Wang, Q.Y. et al. Herpesviral latency–associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 102, 16055–16059 (2005).
Kolb, G. & Kristie, T.M. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J. Virol. 82, 9555–9563 (2008).
Kristie, T.M., Vogel, J.L. & Sears, A.E. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl. Acad. Sci. USA 96, 1229–1233 (1999).
Whitlow, Z. & Kristie, T.M. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J. Virol. 83, 9591–9595 (2009).
Pesola, J.M., Zhu, J., Knipe, D.M. & Coen, D.M. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J. Virol. 79, 14516–14525 (2005).
Sawtell, N.M., Thompson, R.L. & Haas, R.L. Herpes simplex virus DNA synthesis is not a decisive regulatory event in the initiation of lytic viral protein expression in neurons in vivo during primary infection or reactivation from latency. J. Virol. 80, 38–50 (2006).
Oh, J. & Fraser, N.W. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82, 3530–3537 (2008).
Placek, B.J. et al. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 83, 1416–1421 (2009).
Huang, J. et al. p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007).
Wang, J. et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129 (2009).
Acknowledgements
We thank J. Skinner for expert statistical analysis of the experimental data; B. Moss, J. Yewdell, T. Pierson, J. Bennink and A. McBride for critical discussions and comments on this manuscript; members of the Molecular Genetics Section of the Laboratory of Viral Diseases for discussion, advice and technical assistance; the US National Institute of Allergy and Infectious Diseases Bld33 Animal Care Facility staff; N. Fraser (University of Pennsylvania School of Medicine) for HSV-1 strain 17, W. Ruyechan (University at Buffalo, State University of New York) for ICP8–specific sera and X. Chen (University of California at Davis) for MCF7 inducible LSD shRNA cells. These studies were supported by the Laboratory of Viral Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Y.L. performed ChIP, quantitative PCR, immunofluorescence and animal reactivation studies; A.N. and H.P. performed ChIP and quantitative PCR analyses; J.L.V. performed reporter and immunoprecipitation assays; T.M.K. designed the study, performed animal reactivation studies and wrote the paper. J.L.V. and A.N. contributed equally to this study. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The US National Institutes of Health Department of Health and Human Services has filed US patent application No. 61/083,304 and international patent application No. PCT/US2009/051557 for methods of preventing infection by herpes viruses or treating reactivation after latency in a host by inhibitors of the Lsd1 protein.
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–4 and Supplementary Methods (PDF 609 kb)
Rights and permissions
About this article
Cite this article
Liang, Y., Vogel, J., Narayanan, A. et al. Inhibition of the histone demethylase LSD1 blocks α-herpesvirus lytic replication and reactivation from latency. Nat Med 15, 1312–1317 (2009). https://doi.org/10.1038/nm.2051
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2051
This article is cited by
-
Bioinformatics-based analysis of potential candidates chromatin regulators for immune infiltration in osteoarthritis
BMC Musculoskeletal Disorders (2022)
-
Roles and mechanisms of BAP1 deubiquitinase in tumor suppression
Cell Death & Differentiation (2021)
-
The epigenetic implication in coronavirus infection and therapy
Clinical Epigenetics (2020)
-
Epigenetic and epitranscriptomic regulation of viral replication
Nature Reviews Microbiology (2020)
-
Control of viral infections by epigenetic-targeted therapy
Clinical Epigenetics (2019)