Abstract
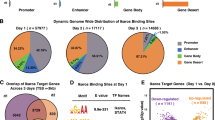
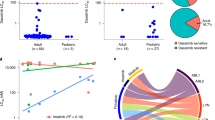
The TLX1 and TLX3 transcription factor oncogenes have a key role in the pathogenesis of T cell acute lymphoblastic leukemia (T-ALL)1,2. Here we used reverse engineering of global transcriptional networks to decipher the oncogenic regulatory circuit controlled by TLX1 and TLX3. This systems biology analysis defined T cell leukemia homeobox 1 (TLX1) and TLX3 as master regulators of an oncogenic transcriptional circuit governing T-ALL. Notably, a network structure analysis of this hierarchical network identified RUNX1 as a key mediator of the T-ALL induced by TLX1 and TLX3 and predicted a tumor-suppressor role for RUNX1 in T cell transformation. Consistent with these results, we identified recurrent somatic loss-of-function mutations in RUNX1 in human T-ALL. Overall, these results place TLX1 and TLX3 at the top of an oncogenic transcriptional network controlling leukemia development, show the power of network analyses to identify key elements in the regulatory circuits governing human cancer and identify RUNX1 as a tumor-suppressor gene in T-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others

References
Ferrando, A.A. et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1, 75–87 (2002).
Aifantis, I., Raetz, E. & Buonamici, S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 8, 380–390 (2008).
Hatano, M., Roberts, C.W., Minden, M., Crist, W.M. & Korsmeyer, S.J. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukemia. Science 253, 79–82 (1991).
Kennedy, M.A. et al. HOX11, a homeobox-containing T-cell oncogene on human chromosome 10q24. Proc. Natl. Acad. Sci. USA 88, 8900–8904 (1991).
Bernard, O.A. et al. A new recurrent and specific cryptic translocation, t(5;14)(q35;q32), is associated with expression of the Hox11L2 gene in T acute lymphoblastic leukemia. Leukemia 15, 1495–1504 (2001).
Van Vlierberghe, P. et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 111, 4668–4680 (2008).
Ferrando, A.A. et al. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet 363, 535–536 (2004).
Su, X.Y. et al. Various types of rearrangements target TLX3 locus in T-cell acute lymphoblastic leukemia. Genes Chromosom. Cancer 41, 243–249 (2004).
Basso, K. et al. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390 (2005).
Margolin, A.A. et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 (suppl. 1), S7 (2006).
De Keersmaecker, K. et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat. Med. 16, 1321–1327 (2010).
Speck, N.A. & Gilliland, D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2, 502–513 (2002).
Song, W.J. et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23, 166–175 (1999).
Osato, M., Yanagida, M., Shigesada, K. & Ito, Y. Point mutations of the RUNx1/AML1 gene in sporadic and familial myeloid leukemias. Int. J. Hematol. 74, 245–251 (2001).
Osato, M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 23, 4284–4296 (2004).
Lefebvre, C. et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6, 377 (2010).
Carro, M.S. et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463, 318–325 (2010).
Preudhomme, C. et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood 113, 5583–5587 (2009).
Owen, C.J. et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 112, 4639–4645 (2008).
Nishimoto, N. et al. T cell acute lymphoblastic leukemia arising from familial platelet disorder. Int. J. Hematol. 92, 194–197 (2010).
Osato, M. et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood 93, 1817–1824 (1999).
Rocquain, J. et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer 10, 401 (2010).
Langabeer, S.E., Gale, R.E., Rollinson, S.J., Morgan, G.J. & Linch, D.C. Mutations of the AML1 gene in acute myeloid leukemia of FAB types M0 and M7. Genes Chromosom. Cancer 34, 24–32 (2002).
Auewarakul, C.U. et al. AML1 mutation and its coexistence with different transcription factor gene families in de novo acute myeloid leukemia (AML): redundancy or synergism. Haematologica 92, 861–862 (2007).
Christiansen, D.H., Andersen, M.K. & Pedersen-Bjergaard, J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood 104, 1474–1481 (2004).
Grossmann, V. et al. Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematol. 96, 1874–1877 (2011).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Margolin, A.A. et al. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc. Natl. Acad. Sci. USA 106, 244–249 (2009).
Fischer, M. et al. MarkUs: a server to navigate sequence-structure-function space. Nucleic Acids Res. 39, W357–W361 (2011).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Acknowledgements
This work was supported by the US National Institutes of Health (grants R01CA120196 and R01CA129382 to A.A.F.; and U24 CA114737 to E.P.), the New York Community Trust (A.A.F.), the Innovative Research Award by the Stand Up to Cancer Foundation (A.A.F.), the Eastern Cooperative Oncology Group tumor bank, the Leukemia and Lymphoma Society Scholar Award (A.A.F.), the Stichting Kinderen Kankervrij (KiKa; grant 2007-012) (J.P.M.) and the Dutch Cancer Society (KWF-EMCR 2006-3500) (J.P.M.). A.P.-G. is a postdoctoral researcher funded by the Rally Foundation. G.D.G. was supported by a Marie Curie International Outgoing fellowship. We thank S. Nimer (Memorial Sloan Kettering Cancer Center) for the pCDNA3 RUNX1 expression vector, D. Zhang (University of California, San Diego) for the pM-CSF-R-luc promoter reporter and the pCMV CBFB plasmids and J. Downing (St. Jude Children's Research Hospital) for the Runx1 knockout mice.
Author information
Authors and Affiliations
Contributions
G.D.G. performed expression and network analysis of human and mouse leukemias, identified RUNX1 mutations in T-ALL and wrote the manuscript; T.P. performed ChIP-chip analysis of TLX1 and TLX3 and the RNA interference experiments; A.P.-G. analyzed the tumor activity of RUNX1; A.A.-I. and M.B. performed network analysis; Z.W.C. performed structure modeling of RUNX1 mutant protein; K.D.K. performed mouse studies; X.S. analyzed ChIP-chip data; L.X. performed mouse studies; E.P., J.R., P.H.W. and J.M.R. provided clinical specimens and correlative data on adult T-ALL samples; J.P.M. contributed clinical specimens and performed microarray analysis; A.C. supervised research; and A.A.F. designed the study, supervised research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–13 and Supplementary Tables 1–11 (PDF 5740 kb)
Rights and permissions
About this article
Cite this article
Della Gatta, G., Palomero, T., Perez-Garcia, A. et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med 18, 436–440 (2012). https://doi.org/10.1038/nm.2610
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2610
This article is cited by
-
Transcription factor network analysis identifies REST/NRSF as an intrinsic regulator of CNS regeneration in mice
Nature Communications (2022)
-
Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases
Communications Biology (2021)
-
Targeted sequencing to identify genetic alterations and prognostic markers in pediatric T-cell acute lymphoblastic leukemia
Scientific Reports (2021)
-
Mutational and functional genetics mapping of chemotherapy resistance mechanisms in relapsed acute lymphoblastic leukemia
Nature Cancer (2020)
-
Master regulator analysis of paragangliomas carrying SDHx, VHL, or MAML3 genetic alterations
BMC Cancer (2019)