Abstract
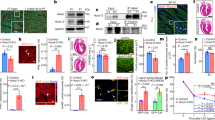
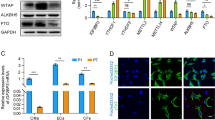
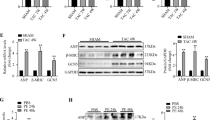
In the adult heart, a variety of stresses induce re-expression of a fetal gene program in association with myocyte hypertrophy and heart failure. Here we show that histone deacetylase-2 (Hdac2) regulates expression of many fetal cardiac isoforms. Hdac2 deficiency or chemical histone deacetylase (HDAC) inhibition prevented the re-expression of fetal genes and attenuated cardiac hypertrophy in hearts exposed to hypertrophic stimuli. Resistance to hypertrophy was associated with increased expression of the gene encoding inositol polyphosphate-5-phosphatase f (Inpp5f) resulting in constitutive activation of glycogen synthase kinase 3β (Gsk3β) via inactivation of thymoma viral proto-oncogene (Akt) and 3-phosphoinositide-dependent protein kinase-1 (Pdk1). In contrast, Hdac2 transgenic mice had augmented hypertrophy associated with inactivated Gsk3β. Chemical inhibition of activated Gsk3β allowed Hdac2-deficient adults to become sensitive to hypertrophic stimulation. These results suggest that Hdac2 is an important molecular target of HDAC inhibitors in the heart and that Hdac2 and Gsk3β are components of a regulatory pathway providing an attractive therapeutic target for the treatment of cardiac hypertrophy and heart failure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jessup, M. & Brozena, S. Heart failure. N. Engl. J. Med. 348, 2007–2018 (2003).
Schrier, R.W. & Abraham, W.T. Hormones and hemodynamics in heart failure. N. Engl. J. Med. 341, 577–585 (1999).
Huss, J.M. & Kelly, D.P. Mitochondrial energy metabolism in heart failure: a question of balance. J. Clin. Invest. 115, 547–555 (2005).
Hoshijima, M. & Chien, K.R. Mixed signals in heart failure: cancer rules. J. Clin. Invest. 109, 849–855 (2002).
Heineke, J. & Molkentin, J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600 (2006).
Izumo, S., Nadal-Ginard, B. & Mahdavi, V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc. Natl. Acad. Sci. USA 85, 339–343 (1988).
Wolffe, A.P. Histone deacetylase: a regulator of transcription. Science 272, 371–372 (1996).
Ekwall, K. Genome-wide analysis of HDAC function. Trends Genet. 21, 608–615 (2005).
Thiagalingam, S. et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. NY Acad. Sci. 983, 84–100 (2003).
McKinsey, T.A. & Olson, E.N. Cardiac histone acetylation–therapeutic opportunities abound. Trends Genet. 20, 206–213 (2004).
Zhang, C.L. et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 (2002).
McKinsey, T.A. & Olson, E.N. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J. Clin. Invest. 115, 538–546 (2005).
Chang, S. et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24, 8467–8476 (2004).
Antos, C.L. et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J. Biol. Chem. 278, 28930–28937 (2003).
Kook, H. et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 112, 863–871 (2003).
Kee, H.J. et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation 113, 51–59 (2006).
Kong, Y. et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation 113, 2579–2588 (2006).
Kook, H. & Epstein, J.A. Hopping to the beat. Hop regulation of cardiac gene expression. Trends Cardiovasc. Med. 13, 261–264 (2003).
Chen, F. et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110, 713–723 (2002).
Shin, C.H. et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110, 725–735 (2002).
Chen, C.S., Weng, S.C., Tseng, P.H., Lin, H.P. & Chen, C.S. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J. Biol. Chem. 280, 38879–38887 (2005).
Yuan, Z.L., Guan, Y.J., Chatterjee, D. & Chin, Y.E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 (2005).
Choi, J.D. et al. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol. Cell. Biol. 25, 5514–5522 (2005).
Minagawa, T., Ijuin, T., Mochizuki, Y. & Takenawa, T. Identification and characterization of a sac domain-containing phosphoinositide 5-phosphatase. J. Biol. Chem. 276, 22011–22015 (2001).
Dorn, G.W. II & Force, T. Protein kinase cascades in the regulation of cardiac hypertrophy. J. Clin. Invest. 115, 527–537 (2005).
Haq, S. et al. Glycogen synthase kinase-3beta is a negative regulator of cardiomyocyte hypertrophy. J. Cell Biol. 151, 117–130 (2000).
Antos, C.L. et al. Activated glycogen synthase-3 beta suppresses cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. USA 99, 907–912 (2002).
Morisco, C. et al. The Akt-glycogen synthase kinase 3beta pathway regulates transcription of atrial natriuretic factor induced by beta-adrenergic receptor stimulation in cardiac myocytes. J. Biol. Chem. 275, 14466–14475 (2000).
Hardt, S.E. & Sadoshima, J. Glycogen synthase kinase-3beta: a novel regulator of cardiac hypertrophy and development. Circ. Res. 90, 1055–1063 (2002).
Zhang, F., Phiel, C.J., Spece, L., Gurvich, N. & Klein, P.S. Inhibitory phosphorylation of glycogen synthase kinase-3 (GSK-3) in response to lithium. Evidence for autoregulation of GSK-3. J. Biol. Chem. 278, 33067–33077 (2003).
Bolden, J.E., Peart, M.J. & Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug. Discov. 9, 769–784 (2006).
Marks, P.A., Richon, V.M., Breslow, R. & Rifkind, R.A. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 13, 477–483 (2001).
Baldinu, P. et al. Identification of a novel candidate gene, CASC2, in a region of common allelic loss at chromosome 10q26 in human endometrial cancer. Hum. Mutat. 23, 318–326 (2004).
Peiffer-Schneider, S. et al. Mapping an endometrial cancer tumor suppressor gene at 10q25 and development of a bacterial clone contig for the consensus deletion interval. Genomics 52, 9–16 (1998).
Marks, P.A. & Dokmanovic, M. Histone deacetylase inhibitors: discovery and development as anticancer agents. Expert Opin. Investig. Drugs 14, 1497–1511 (2005).
Skov, S. et al. Cancer cells become susceptible to natural killer cell killing after exposure to histone deacetylase inhibitors due to glycogen synthase kinase-3-dependent expression of MHC class I-related chain a and B. Cancer Res. 65, 11136–11145 (2005).
Johnson, C.A. & Turner, B.M. Histone deacetylases: complex transducers of nuclear signals. Semin. Cell Dev. Biol. 10, 179–188 (1999).
Rockman, H.A. et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 88, 8277–8281 (1991).
Zhou, R., Pickup, S., Glickson, J.D., Scott, C.H. & Ferrari, V.A. Assessment of global and regional myocardial function in the mouse using cine and tagged MRI. Magn. Reson. Med. 49, 760–764 (2003).
Matsui, T. et al. Adenoviral gene transfer of activated phosphatidylinositol 3′-kinase and Akt inhibits apoptosis of hypoxic cardiomyocytes in vitro. Circulation 100, 2373–2379 (1999).
Wang, Q. et al. Protein kinase B/Akt participates in GLUT4 translocation by insulin in L6 myoblasts. Mol. Cell. Biol. 19, 4008–4018 (1999).
Acknowledgements
We thank J. Tobias for his help with microarray data analysis; A. Granger and M. Levin for assistance with cardiac myocyte isolation; K.J. Duffy for his help with analysis of echocardiography data; R. Zhou for assistance with magnetic resonance imaging; and T. Force (Thomas Jefferson University) for dnAkt and caAkt adenovirus and constructs, and for advice with the manuscript. This work was supported by the US National Institutes of Health (RO1 HL071546 to J.A.E.). J.A.E. holds the W.W. Smith Endowed Chair for Cardiovascular Research at the University of Pennsylvania. C.M.T. is supported by an American Heart Association postdoctoral fellowship.
Author information
Authors and Affiliations
Contributions
C.M.T. contributed significantly to the writing of the manuscript. M.Z. performed histological sectioning of embryo and heart tissue. W.Z. assisted with siRNA experiments. T.W. performed TAC surgery. T.F., M.G., P.R.N. and W.W. created the Hdac2 gene-trap ES line. V.A.F. carried out echocardiography and MRI studies. C.S.A. helped with PI3K activity experiments and provided advice related to PI3K signaling. P.J.G. was instrumental during early stages of the project and initiated Hdac2 expression studies. J.A.E. conceived, designed and directed the study, supervised C.M.T., Y.L., Z.Y., M.Z., T.W. and W.Z., and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.L. declares that he is presently employed by Novartis Pharmaceuticals, though he did not work for Novartis at the time that the work was performed in the Epstein lab. V.F. declares that he receives grant support from GlaxoSmithKline, Inc. Other authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Transgenic over-expression of Hdac1 or Hdac3 does not affect Inpp5f expression. (PDF 270 kb)
Supplementary Fig. 2
Regulation of Inpp5f expression in H9c2 myocytes. (PDF 262 kb)
Supplementary Fig. 3
Loss of Hdac2 does not affect activity of Pkc, PKA, or Ilk. (PDF 569 kb)
Supplementary Table 1
Genotypes of Hdac2+/− intercrosses. (PDF 48 kb)
Supplementary Table 2
Proliferation of myocardial cells. (PDF 49 kb)
Supplementary Table 3
Measurements of cardiac hypertrophy at P60-80. (PDF 68 kb)
Rights and permissions
About this article
Cite this article
Trivedi, C., Luo, Y., Yin, Z. et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3β activity. Nat Med 13, 324–331 (2007). https://doi.org/10.1038/nm1552
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1552
This article is cited by
-
HDAC5 inhibition attenuates ventricular remodeling and cardiac dysfunction
Orphanet Journal of Rare Diseases (2023)
-
Pregnancy-induced physiological hypertrophic preconditioning attenuates pathological myocardial hypertrophy by activation of FoxO3a
Cellular and Molecular Life Sciences (2023)
-
Posttranslational Modifications: Emerging Prospects for Cardiac Regeneration Therapy
Journal of Cardiovascular Translational Research (2022)
-
DNA methylation abnormalities of imprinted genes in congenital heart disease: a pilot study
BMC Medical Genomics (2021)
-
Ketogenic diets inhibit mitochondrial biogenesis and induce cardiac fibrosis
Signal Transduction and Targeted Therapy (2021)