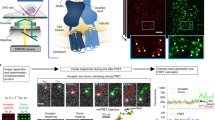
Abstract
Cell-surface proteins are important in cell-cell communication. They assemble into heterocomplexes that include different receptors and effectors. Elucidation and manipulation of such protein complexes offers new therapeutic possibilities. We describe a methodology combining time-resolved fluorescence resonance energy transfer (FRET) with snap-tag technology to quantitatively analyze protein-protein interactions at the surface of living cells, in a high throughput–compatible format. Using this approach, we examined whether G protein–coupled receptors (GPCRs) are monomers or assemble into dimers or larger oligomers—a matter of intense debate. We obtained evidence for the oligomeric state of both class A and class C GPCRs. We also observed different quaternary structure of GPCRs for the neurotransmitters glutamate and γ-aminobutyric acid (GABA): whereas metabotropic glutamate receptors assembled into strict dimers, the GABAB receptors spontaneously formed dimers of heterodimers, offering a way to modulate G-protein coupling efficacy. This approach will be useful in systematic analysis of cell-surface protein interaction in living cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bockaert, J. & Pin, J.P. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 18, 1723–1729 (1999).
Bayburt, T.H., Leitz, A.J., Xie, G., Oprian, D.D. & Sligar, S.G. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 282, 14875–14881 (2007).
Chabre, M. & le Maire, M. Monomeric G-protein-coupled receptor as a functional unit. Biochemistry 44, 9395–9403 (2005).
Ernst, O.P., Gramse, V., Kolbe, M., Hofmann, K.P. & Heck, M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc. Natl. Acad. Sci. USA 104, 10859–10864 (2007).
Whorton, M.R. et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA 104, 7682–7687 (2007).
Bouvier, M. Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2, 274–286 (2001).
Milligan, G. G-protein-coupled receptor heterodimers: pharmacology, function and relevance to drug discovery. Drug Discov. Today 11, 541–549 (2006).
Salahpour, A. & Masri, B. Experimental challenge to a 'rigorous' BRET analysis of GPCR oligomerization. Nat. Methods 4, 599–600 (2007).
James, J.R., Oliveira, M.I., Carmo, A.M., Iaboni, A. & Davis, S.J. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat. Methods 3, 1001–1006 (2006).
Meyer, B.H. et al. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc. Natl. Acad. Sci. USA 103, 2138–2143 (2006).
Pfleger, K.D. & Eidne, K.A. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods 3, 165–174 (2006).
Selvin, P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7, 730–734 (2000).
Bazin, H., Trinquet, E. & Mathis, G. Time resolved amplification of cryptate emission: a versatile technology to trace biomolecular interactions. J. Biotechnol. 82, 233–250 (2002).
Kniazeff, J. et al. Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol. 11, 706–713 (2004).
Maurel, D. et al. Cell surface detection of membrane protein interaction with homogeneous time-resolved fluorescence resonance energy transfer technology. Anal. Biochem. 329, 253–262 (2004).
McVey, M. et al. Monitoring receptor oligomerization using time-resolved fluorescence resonance energy transfer and bioluminescence resonance energy transfer. The human delta-opioid receptor displays constitutive oligomerization at the cell surface, which is not regulated by receptor occupancy. J. Biol. Chem. 276, 14092–14099 (2001).
Keppler, A., Pick, H., Arrivoli, C., Vogel, H. & Johnsson, K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA 101, 9955–9959 (2004).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Pin, J.-P. Allosteric functioning of dimeric Class C G-protein coupled receptors. FEBS J. 272, 2947–2955 (2005).
Brock, C., Boudier, L., Maurel, D., Blahos, J. & Pin, J.P. Assembly-dependent surface targeting of the heterodimeric GABAB Receptor is controlled by COPI but not 14–3-3. Mol. Biol. Cell 16, 5572–5578 (2005).
Margeta-Mitrovic, M., Jan, Y.N. & Jan, L.Y. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27, 97–106 (2000).
Brock, C. et al. Activation of a dimeric metabotropic glutamate receptor by inter-subunit rearrangement. J. Biol. Chem. 282, 33000–33008 (2007).
Bischoff, S. et al. Spatial distribution of GABA(B)R1 receptor mRNA and binding sites in the rat brain. J. Comp. Neurol. 412, 1–16 (1999).
Kaupmann, K. et al. Expression cloning of GABA(B) receptors uncovers similarity to metabotropic glutamate receptors. Nature 386, 239–246 (1997).
Selvin, P.R. & Hearst, J.E. Luminescence energy transfer using a terbium chelate: improvements on fluorescence energy transfer. Proc. Natl. Acad. Sci. USA 91, 10024–10028 (1994).
Villemure, J.F. et al. Subcellular distribution of GABA(B) receptor homo- and hetero-dimers. Biochem. J. 388, 47–55 (2005).
Lopez-Gimenez, J.F., Canals, M., Pediani, J.D. & Milligan, G. The alpha1b-adrenoceptor exists as a higher-order oligomer: effective oligomerization is required for receptor maturation, surface delivery, and function. Mol. Pharmacol. 71, 1015–1029 (2007).
Damian, M., Martin, A., Mesnier, D., Pin, J.P. & Banères, J.L. Asymmetric conformational changes in a GPCR dimer controlled by G-proteins. EMBO J. 25, 5693–5702 (2006).
Hlavackova, V. et al. Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 24, 499–509 (2005).
White, J.F. et al. Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc. Natl. Acad. Sci. USA 104, 12199–12204 (2007).
Acknowledgements
We thank C. Vol for expert assistance with the GPCR functional assays; F. Maurin (Cisbio) for his participation in the synthesis of BG derivatives; and K. Johnsson (Ecole Polytechnique Fédérale de Lausanne) for his support to this project, his critical reading of the manuscript and for providing us with snap-tag tools. We also thank P. Rondard and R. Jockers for their comments on the manuscript. Confocal imaging was performed at the Centre de Ressources en Imagerie Cellulaire, Montpellier, with the help of N. Lautredou. This work was made possible thanks to the screening facilities of the Institut Fédératif de Recherche 3. This work was supported by CNRS, INSERM, Cisbio and by grants from the French Ministry of Research, Action Concertée Incitative “Biologie Cellulaire Moléculaire et Structurale” (ACI-BCMS 328), the Agence Nationale de la Recherche (ANR-05-PRIB-02502, ANR-BLAN06-3_135092 and ANR-05-NEUR-035) and by an unrestricted grant from Senomyx. D.M. was supported by the Association pour le Dévelopement de l'Enseignement et de la Recherche Languedoc Roussillon and C.B. by the Fondation Recherche Médicale.
Author information
Authors and Affiliations
Contributions
D.M. and L.C.A. executed most of the experiments and participated in the writing of the manuscript; C.B. developed the system to control subunit composition and performed the confocal experiments; M.L.R. performed the experiments with mGlu1 receptor and the initial experiments with class A receptors; E.B. and H.B. synthesized the BG derivatives; M.A. participated in the BRET experiments; N.T. and E.T. supervised the work at Cisbio; T.D. and L.P. supervised some aspects of the work at the IGF; and J.P.P. supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.B., H.B., N.T. and E.T. are employees of Cisbio, which develops products combining its htrf technology with Covalys SNAP-tag protein labeling system, and therefore may gain financially through publication of this paper. Part of the work of CNRS UMR 5203 has been financially supported by Cisbio and by an unrestricted grant from Senomyx.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Methods (PDF 409 kb)
Rights and permissions
About this article
Cite this article
Maurel, D., Comps-Agrar, L., Brock, C. et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat Methods 5, 561–567 (2008). https://doi.org/10.1038/nmeth.1213
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1213
This article is cited by
-
Filamin A organizes γ‑aminobutyric acid type B receptors at the plasma membrane
Nature Communications (2023)
-
Mesenchymal stem cells transfer mitochondria to allogeneic Tregs in an HLA-dependent manner improving their immunosuppressive activity
Nature Communications (2022)
-
Biased signaling due to oligomerization of the G protein-coupled platelet-activating factor receptor
Nature Communications (2022)
-
Determination of G-protein–coupled receptor oligomerization by molecular brightness analyses in single cells
Nature Protocols (2021)
-
Single-molecule FRET imaging of GPCR dimers in living cells
Nature Methods (2021)