Abstract
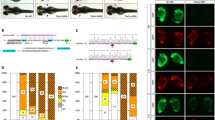
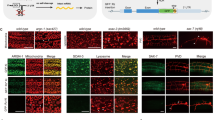
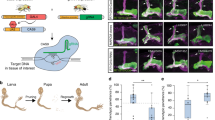
We describe a conditional in vivo protein-trap mutagenesis system that reveals spatiotemporal protein expression dynamics and can be used to assess gene function in the vertebrate Danio rerio. Integration of pGBT-RP2.1 (RP2), a gene-breaking transposon containing a protein trap, efficiently disrupts gene expression with >97% knockdown of normal transcript amounts and simultaneously reports protein expression for each locus. The mutant alleles are revertible in somatic tissues via Cre recombinase or splice-site-blocking morpholinos and are thus to our knowledge the first systematic conditional mutant alleles outside the mouse model. We report a collection of 350 zebrafish lines that include diverse molecular loci. RP2 integrations reveal the complexity of genomic architecture and gene function in a living organism and can provide information on protein subcellular localization. The RP2 mutagenesis system is a step toward a unified 'codex' of protein expression and direct functional annotation of the vertebrate genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Stevens, C.W. The evolution of vertebrate opioid receptors. Front. Biosci. 14, 1247–1269 (2009).
Huxley-Jones, J., Robertson, D.L. & Boot-Handford, R.P. On the origins of the extracellular matrix in vertebrates. Matrix Biol. 26, 2–11 (2007).
Sauka-Spengler, T. & Bronner-Fraser, M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis 46, 673–682 (2008).
Balciunas, D. et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 (2006).
Urasaki, A., Morvan, G. & Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174, 639–649 (2006).
Sivasubbu, S. et al. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech. Dev. 123, 513–529 (2006).
Petzold, A.M. et al. Nicotine response genetics in the zebrafish. Proc. Natl. Acad. Sci. USA 106, 18662–18667 (2009).
Branda, C.S. & Dymecki, S.M. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 (2004).
Parinov, S., Kondrichin, I., Korzh, V. & Emelyanov, A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 (2004).
Balciunas, D. et al. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5, 62 (2004).
Kawakami, K. et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144 (2004).
Munoz-Sanjuan, I., Simandl, B.K., Fallon, J.F. & Nathans, J. Expression of chicken fibroblast growth factor homologous factor (FHF)-1 and of differentially spliced isoforms of FHF-2 during development and involvement of FHF-2 in chicken limb development. Development 126, 409–421 (1999).
Holland, P.W. Beyond the Hox: how widespread is homeobox gene clustering? J. Anat. 199, 13–23 (2001).
Coulombe, Y. et al. Multiple promoters and alternative splicing: Hoxa5 transcriptional complexity in the mouse embryo. PLoS ONE 5, e10600 (2010).
Hadrys, T., Prince, V., Hunter, M., Baker, R. & Rinkwitz, S. Comparative genomic analysis of vertebrate Hox3 and Hox4 genes. J. Exp. Zoolog. B Mol. Dev. Evol. 302, 147–164 (2004).
Chen, J.N. et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development 123, 293–302 (1996).
Sehnert, A.J. et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 31, 106–110 (2002).
Hirata, H. et al. Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development 134, 2771–2781 (2007).
Schoenauer, R. et al. Myomesin 3, a novel structural component of the M-band in striated muscle. J. Mol. Biol. 376, 338–351 (2008).
Klee, E.W. The zebrafish secretome. Zebrafish 5, 131–138 (2008).
Carney, T.J. et al. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet. 6, e1000907 (2010).
Gautier, P., Naranjo-Golborne, C., Taylor, M.S., Jackson, I.J. & Smyth, I. Expression of the fras1/frem gene family during zebrafish development and fin morphogenesis. Dev. Dyn. 237, 3295–3304 (2008).
van Eeden, F.J. et al. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development 123, 255–262 (1996).
Friedel, R.H. & Soriano, P. in Methods in Enzymology Vol. 477 (eds., Wassarman, P.M. & Soriano, P.M.) 243–269 (Academic Press, 2010).
Aleksic, J., Lazic, R., Müller, I., Russell, S.R. & Adryan, B. Biases in Drosophila melanogaster protein trap screens. BMC Genomics 10, 249 (2009).
Buszczak, M. et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 (2007).
Morin, X., Daneman, R., Zavortink, M. & Chia, W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15050–15055 (2001).
Nord, A.S. et al. Modeling insertional mutagenesis using gene length and expression in murine embryonic stem cells. PLoS ONE 2, e617 (2007).
Skarnes, W.C. et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 36, 543–544 (2004).
Cormack, B.P., Valdivia, R.H. & Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 (1996).
Gibbs, P. & Schmale, M. GFP as a genetic marker scorable throughout the life cycle of transgenic zebra fish. Mar. Biotechnol. 2, 107–125 (2000).
Campbell, R.E. et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882 (2002).
Petzold, A.M. et al. SCORE imaging: specimen in a corrected optical rotational enclosure. Zebrafish 7, 149–154 (2010).
Clark, K.J., Geurts, A.M., Bell, J.B. & Hackett, P.B. Transposon vectors for gene-trap insertional mutagenesis in vertebrates. Genesis 39, 225–233 (2004).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Bill, B.R., Petzold, A.M., Clark, K.J., Schimmenti, L.A. & Ekker, S.C. A primer for morpholino use in zebrafish. Zebrafish 6, 69–77 (2009).
Acknowledgements
The US National Institute on Drug Abuse (DA14546), US National Institute of General Medical Sciences (GM63904), National Institute of Diabetes and Digestive and Kidney Diseases (F30DK083219 and P30DK084567) and the Mayo Foundation provided funding for this research. S.S. and V.S. acknowledge funding support from the Council of Scientific and Industrial Research (grant FAC002), India. We thank members of the Center for Genome Engineering at the University of Minnesota for providing collaborative discussion and resources, the staff of the Zebrafish Core Facility at the Mayo Clinic for providing zebrafish care, D. Argue for programming the zfishbook website, E. Klee for bioinformatic analyses of the zebrafish transcriptome used in theoretical design and testing of the RP2 system, A. Person for assistance in injection of fluorescently labeled dextrans, and InSciEd Out participants and Summer Undergraduate Research Fellow, N. Boczek, for imaging RFP-expressing fish.
Author information
Authors and Affiliations
Contributions
K.J.C., D.B., S.S. and S.C.E. designed the experiments. D.B. built R14, R15, R16, RP2 and pEF1a-cd99l2-RFP vectors. S.S. built pX. J.N. made the pEF1a-RFP vector. D.B. piloted the production of R14, R15 and R16 lines. K.J.C. trained and managed the team that produced >300 RP2 lines and molecularly characterized lines other than GBT0031, GBT0039 and GBT0043 (D.B.). M.D.U., T.M.G., A.L.N., K.J.S. and K.J.C. identified and propagated mRFP-expressing lines. A.M.P., M.D.U. and K.J.C. photographed mRFP expression patterns. K.J.S. and K.J.C. molecularly characterized lines by 5′ rapid amplification of cDNA ends, inverse PCR and quantitative PCR. T.M.G., K.J.C. and D.B. characterized the reversion of GBT0031 by Cre mRNA and morpholino injection. D.B. injected fluorescently conjugated dextran into GBT0046 and imaged its uptake. J.N. imaged GBT0043 fish and together with K.J.C. injected fish with EF1a-RFP and EF1a-cd99l2-RFP vectors. K.J.C. imaged the injected fish. S.E.W. analyzed the GBT0156/frasI phenotype and reversion. V.M.B. conducted in situ hybridizations. Y.D. identified GBT0348 in a screen performed in X.X.'s laboratory and contributed quantitative PCR data for three more RP2 lines. H.-M.P. tested the GBT protocol in M.H.'s lab and produced several independent lines, including the initial assessment and imaging of GBT0040. A.P., V.S. and S.S. produced and analyzed 3′ exon trap data for Supplementary Figure 2. K.J.C. and S.C.E. were primary authors of the text.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Tables 1–2 and Supplementary Note (PDF 4469 kb)
Rights and permissions
About this article
Cite this article
Clark, K., Balciunas, D., Pogoda, HM. et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods 8, 506–512 (2011). https://doi.org/10.1038/nmeth.1606
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1606
This article is cited by
-
Multiple pkd and piezo gene family members are required for atrioventricular valve formation
Nature Communications (2023)
-
Deficit of homozygosity among 1.52 million individuals and genetic causes of recessive lethality
Nature Communications (2023)
-
Loss of Neuropilin2a/b or Sema3fa alters olfactory sensory axon dynamics and protoglomerular targeting
Neural Development (2022)
-
Protein trap: a new Swiss army knife for geneticists?
Molecular Biology Reports (2020)
-
Zebrafish Embryonic Slow Muscle Is a Rapid System for Genetic Analysis of Sarcomere Organization by CRISPR/Cas9, but Not NgAgo
Marine Biotechnology (2018)