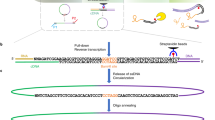
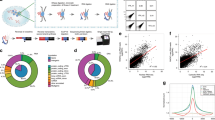
Abstract
Identifying the chromosomal targets of transcription factors is important for reconstructing the transcriptional regulatory networks underlying global gene expression programs. We have developed an unbiased genomic method called sequence tag analysis of genomic enrichment (STAGE) to identify the direct binding targets of transcription factors in vivo. STAGE is based on high-throughput sequencing of concatemerized tags derived from target DNA enriched by chromatin immunoprecipitation. We first used STAGE in yeast to confirm that RNA polymerase III genes are the most prominent targets of the TATA-box binding protein. We optimized the STAGE protocol and developed analysis methods to allow the identification of transcription factor targets in human cells. We used STAGE to identify several previously unknown binding targets of human transcription factor E2F4 that we independently validated by promoter-specific PCR and microarray hybridization. STAGE provides a means of identifying the chromosomal targets of DNA-associated proteins in any sequenced genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Pollack, J.R. & Iyer, V.R. Characterizing the physical genome. Nat. Genet. 32 (Suppl.), 515–521 (2002).
Lee, T.I. et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804 (2002).
Yu, H., Luscombe, N.M., Qian, J. & Gerstein, M. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 19, 422–427 (2003).
Phimister, B. Getting hip to the chip. Nat. Genet. 18, 195–197 (1998).
Iyer, V.R. et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409, 533–538 (2001).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Ren, B. et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245–256 (2002).
Weinmann, A.S., Yan, P.S., Oberley, M.J., Huang, T.H. & Farnham, P.J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16, 235–244 (2002).
Martone, R. et al. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. USA 100, 12247–12252 (2003).
Cawley, S. et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509 (2004).
Weinmann, A.S., Bartley, S.M., Zhang, T., Zhang, M.Q. & Farnham, P.J. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21, 6820–6832 (2001).
Velculescu, V.E., Zhang, L., Vogelstein, B. & Kinzler, K.W. Serial analysis of gene expression. Science 270, 484–487 (1995).
Velculescu, V.E., Vogelstein, B. & Kinzler, K.W. Analysing uncharted transcriptomes with SAGE. Trends Genet. 16, 423–425 (2000).
Roberts, D.N., Stewart, A.J., Huff, J.T. & Cairns, B.R. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA 100, 14695–14700 (2003).
Kim, J. & Iyer, V.R. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol. Cell. Biol. 24, 8104–8112 (2004).
Hahn, J.S., Hu, Z., Thiele, D.J. & Iyer, V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24, 5249–5256 (2004).
Cam, H. & Dynlacht, B.D. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3, 311–316 (2003).
Pruitt, K.D., Tatusova, T. & Maglott, D.R. NCBI Reference Sequence project: update and current status. Nucleic Acids Res. 31, 34–37 (2003).
Karolchik, D. et al. The UCSC Genome Browser Database. Nucleic Acids Res. 31, 51–54 (2003).
Odom, D.T. et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 303, 1378–1381 (2004).
Saha, S. et al. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20, 508–512 (2002).
Matsumura, H. et al. Gene expression analysis of plant host-pathogen interactions by SuperSAGE. Proc. Natl. Acad. Sci. USA 100, 15718–15723 (2003).
Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 (2003).
Hild, M. et al. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 5, R3 (2003).
Kuras, L. & Struhl, K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399, 609–613 (1999).
Iyer, V.R. in DNA Microarrays: A Molecular Cloning Manual (eds. D. Bowtell & J. Sambrook) 453–463 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2003).
Killion, P.J., Sherlock, G. & Iyer, V.R. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD). BMC Bioinformatics 4, 32 (2003).
Acknowledgements
We thank K. Struhl for the HA-tagged TBP strain, P. Killion for assistance with the microarray database and T. Hart and members of the Iyer lab for assistance with microarray production. This work was supported in part by a grant from the Texas State Higher Education Coordinating Board, a US Department of Defense Breast Cancer Idea Award and a National Science Foundation Information Technology Research (ITR) grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Summary of STAGE tags sequenced (PDF 12 kb)
Supplementary Table 2
Gene scores from E2F4 STAGE and SubSTAGE (PDF 114 kb)
Supplementary Table 3
Human E2F4 targets predicted by STAGE (between 10 kb and 6 kb upstream of transcription start site) (PDF 92 kb)
Supplementary Table 4
Human E2F4 targets predicted by STAGE (between 6 kb and 2 kb upstream of transcription start site) (PDF 80 kb)
Supplementary Table 5
Human E2F4 targets predicted by STAGE (1st intron) (PDF 63 kb)
Supplementary Table 6
Primer pairs for ChIP-PCR verification (PDF 92 kb)
Rights and permissions
About this article
Cite this article
Kim, J., Bhinge, A., Morgan, X. et al. Mapping DNA-protein interactions in large genomes by sequence tag analysis of genomic enrichment. Nat Methods 2, 47–53 (2005). https://doi.org/10.1038/nmeth726
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth726
This article is cited by
-
In silico regulatory analysis for exploring human disease progression
Biology Direct (2008)
-
Identification of novel androgen receptor target genes in prostate cancer
Molecular Cancer (2007)
-
Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing
Nature Methods (2007)
-
Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project
Nature (2007)