Abstract
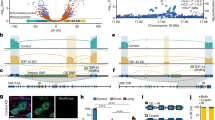
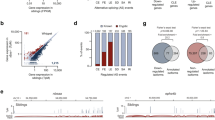
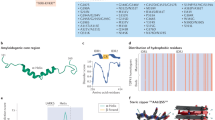
TDP-43 is a predominantly nuclear RNA-binding protein that forms inclusion bodies in frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). The mRNA targets of TDP-43 in the human brain and its role in RNA processing are largely unknown. Using individual nucleotide-resolution ultraviolet cross-linking and immunoprecipitation (iCLIP), we found that TDP-43 preferentially bound long clusters of UG-rich sequences in vivo. Analysis of RNA binding by TDP-43 in brains from subjects with FTLD revealed that the greatest increases in binding were to the MALAT1 and NEAT1 noncoding RNAs. We also found that binding of TDP-43 to pre-mRNAs influenced alternative splicing in a similar position-dependent manner to Nova proteins. In addition, we identified unusually long clusters of TDP-43 binding at deep intronic positions downstream of silenced exons. A substantial proportion of alternative mRNA isoforms regulated by TDP-43 encode proteins that regulate neuronal development or have been implicated in neurological diseases, highlighting the importance of TDP-43 for the regulation of splicing in the brain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Buratti, E. & Baralle, F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 13, 867–878 (2008).
Kuo, P.H., Doudeva, L.G., Wang, Y.T., Shen, C.K. & Yuan, H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 37, 1799–1808 (2009).
Buratti, E. & Baralle, F.E. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 (2001).
Sreedharan, J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008).
Lagier-Tourenne, C., Polymenidou, M. & Cleveland, D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64 (2010).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Igaz, L.M. et al. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 284, 8516–8524 (2009).
Johnson, B.S., McCaffery, J.M., Lindquist, S. & Gitler, A.D. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 105, 6439–6444 (2008).
Voigt, A. et al. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS ONE 5, e12247 (2010).
König, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 (2010).
Clemson, C.M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Cleveland, D.W. & Rothstein, J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2, 806–819 (2001).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003).
Ayala, Y.M. et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288 (2011).
Polymenidou, M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. advance online publication, 10.1038/nn.2779 (27 February 2011).
Sephton, C.F. et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 286, 1204–1215 (2011).
Strong, M.J. et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 35, 320–327 (2007).
Ayala, Y.M., Misteli, T. & Baralle, F.E. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc. Natl. Acad. Sci. USA 105, 3785–3789 (2008).
Buratti, E. et al. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 277, 2268–2281 (2010).
Kametani, F. et al. Identification of casein kinase-1 phosphorylation sites on TDP-43. Biochem. Biophys. Res. Commun. 382, 405–409 (2009).
Fiesel, F.C. et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221 (2010).
O'Connor, L. et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17, 384–395 (1998).
Cléry, A., Blatter, M. & Allain, F.H. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 18, 290–298 (2008).
Witten, J.T. & Ule, J. Understanding splicing regulation through RNA splicing maps. Trends Genet. published online, doi:10.1016/j.tig.2010.12.001 (11 January 2011).
Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 (2003).
Hung, L.H. et al. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA 14, 284–296 (2008).
Rockman, M.V. & Wray, G.A. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 19, 1991–2004 (2002).
Kashi, Y. & King, D.G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 22, 253–259 (2006).
Legendre, M., Pochet, N., Pak, T. & Verstrepen, K.J. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 17, 1787–1796 (2007).
Wu, L.S. et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 48, 56–62 (2010).
Matter, C., Pribadi, M., Liu, X. & Trachtenberg, J.T. Delta-catenin is required for the maintenance of neural structure and function in mature cortex in vivo. Neuron 64, 320–327 (2009).
Ishiyama, N. et al. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 141, 117–128 (2010).
Kouchi, Z. et al. p120 catenin recruits cadherins to gamma-secretase and inhibits production of Aβ peptide. J. Biol. Chem. 284, 1954–1961 (2009).
Yang, Q. et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323, 124–127 (2009).
Ness, J.M. et al. Selective involvement of BH3-only Bcl-2 family members Bim and Bad in neonatal hypoxia-ischemia. Brain Res. 1099, 150–159 (2006).
Becker, E.B. & Bonni, A. Pin1 in neuronal apoptosis. Cell Cycle 6, 1332–1335 (2007).
Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010).
Wang, Z. et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 8, e1000530 (2010).
Acknowledgements
We thank K. Zarnack for advice and comments on the manuscript, F. Baralle, Y. Ayalla and A.D. Ambrogio for HeLa knockdown RNA, and J. Hadfield and the genomic team at CRI for Illumina sequencing. This work was supported by the European Research Council (206726-CLIP), the MRC, Slovenian Research Agency (P2-0209, J2-2197, L2-1112, Z7-3665), Wellcome Trust and MRC Strategic Grant Award (089701/Z/09/Z), Motor Neuron Disease Association, Heaton-Ellis Trust and Psychiatry Research Trust.
Author information
Authors and Affiliations
Contributions
J.R.T. carried out TDP-43 iCLIP, microarray and PCR experiments. M.B. carried out CELF2 iCLIP. T.C. and G.R. mapped the iCLIP sequence reads to genome, evaluated random barcodes, determined cross-link clusters and annotated the data. T.C. analyzed the reproducibility, sequence and positioning of TDP-43 cross-link sites and performed gene ontology analysis. B.R., A.L.N. and V.Ž. prepared RNA from knockdown cells and brain tissue. T.H. selected, sampled and analyzed the brain samples. M.C. and M.K. analyzed splice-junction microarray data and generated the RNA splicing map. R.P. prepared the embryonic stem cells. S.C., C.E.S., B.Z., J.K. and J.U. supervised the project. J.R.T., T.C., B.R., J.K., C.E.S. and J.U. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Tables 1–4 (PDF 5584 kb)
Rights and permissions
About this article
Cite this article
Tollervey, J., Curk, T., Rogelj, B. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14, 452–458 (2011). https://doi.org/10.1038/nn.2778
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2778
This article is cited by
-
Stathmin-2 loss leads to neurofilament-dependent axonal collapse driving motor and sensory denervation
Nature Neuroscience (2024)
-
Genome-wide analyses identify NEAT1 as genetic modifier of age at onset of amyotrophic lateral sclerosis
Molecular Neurodegeneration (2023)
-
Specific vulnerability of iPSC-derived motor neurons with TDP-43 gene mutation to oxidative stress
Molecular Brain (2023)
-
Aggregation-prone TDP-43 sequesters and drives pathological transitions of free nuclear TDP-43
Cellular and Molecular Life Sciences (2023)
-
Slow motor neurons resist pathological TDP-43 and mediate motor recovery in the rNLS8 model of amyotrophic lateral sclerosis
Acta Neuropathologica Communications (2022)