Abstract
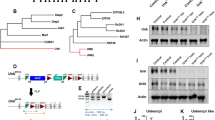
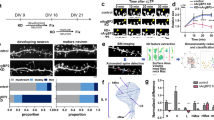
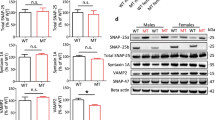
Oligophrenin-1 regulates dendritic spine morphology in the brain. Mutations in the oligophrenin-1 gene (OPHN1) cause intellectual disability. We discovered a previously unknown partner of oligophrenin-1, Rev-erbα, a nuclear receptor that represses the transcription of circadian oscillators. We found that oligophrenin-1 interacts with Rev-erbα in the mouse brain, causing it to locate to dendrites, reducing its repressor activity and protecting it from degradation. Our results indicate the presence of a circadian oscillator in the hippocampus, involving the clock gene Bmal1 (also known as Arntl), that is modulated by Rev-erbα and requires oligophrenin-1 for normal oscillation. We also found that synaptic activity induced Rev-erbα localization to dendrites and spines, a process that is mediated by AMPA receptor activation and requires oligophrenin-1. Our data reveal new interactions between synaptic activity and circadian oscillators, and delineate a new means of communication between nucleus and synapse that may provide insight into normal plasticity and the etiology of intellectual disability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
11 September 2011
In the version of this article initially published online, affiliation 4 was misnumbered as 5, 5 was misnumbered as 6 and 6 was misnumbered as 4. The error has been corrected for the print, PDF and HTML versions of this article.
References
Gécz, J., Shoubridge, C. & Corbett, M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 25, 308–316 (2009).
Kaufman, L., Ayub, M. & Vincent, J.B . The genetic basis of non-syndromic intellectual disability: a review. J. Neurodev. Disord. 2, 182–209 (2010).
Ropers, H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 11, 161–187 (2010).
Ropers, H.H. & Hamel, B.C. X-linked mental retardation. Nat. Rev. Genet. 6, 46–57 (2005).
Fauchereau, F. et al. The RhoGAP activity of OPHN1, a new F-actin-binding protein, is negatively controlled by its amino-terminal domain. Mol. Cell. Neurosci. 23, 574–586 (2003).
Govek, E.E. et al. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat. Neurosci. 7, 364–372 (2004).
Khelfaoui, M. et al. Loss of X-linked mental retardation gene oligophrenin1 in mice impairs spatial memory and leads to ventricular enlargement and dendritic spine immaturity. J. Neurosci. 27, 9439–9450 (2007).
Billuart, P. et al. Oligophrenin 1 encodes a rho-GAP protein involved in X-linked mental retardation. Nature 392, 923–926 (1998).
Tentler, D. et al. Deletion including the oligophrenin-1 gene associated with enlarged cerebral ventricles, cerebellar hypoplasia, seizures and ataxia. Eur. J. Hum. Genet. 7, 541–548 (1999).
Philip, N. et al. Mutations in the oligophrenin-1 gene (OPHN1) cause X linked congenital cerebellar hypoplasia. J. Med. Genet. 40, 441–446 (2003).
Bergmann, C. et al. Oligophrenin 1 (OPHN1) gene mutation causes syndromic X-linked mental retardation with epilepsy, rostral ventricular enlargement and cerebellar hypoplasia. Brain 126, 1537–1544 (2003).
Nakano-Kobayashi, A., Kasri, N.N., Newey, S.E. & Van Aelst, L. The Rho-linked mental retardation protein OPHN1 controls synaptic vesicle endocytosis via endophilin A1. Curr. Biol. 19, 1133–1139 (2009).
Xiao, B., Tu, J.C. & Worley, P.F. Homer: a link between neural activity and glutamate receptor function. Curr. Opin. Neurobiol. 10, 370–374 (2000).
Fagni, L., Chavis, P., Ango, F. & Bockaert, J. Complex interactions between mGluRs, intracellular Ca2+ stores and ion channels in neurons. Trends Neurosci. 23, 80–88 (2000).
Sala, C. et al. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31, 115–130 (2001).
Khelfaoui, M. et al. Inhibition of RhoA pathway rescues the endocytosis defects in Oligophrenin1 mouse model of mental retardation. Hum. Mol. Genet. 18, 2575–2583 (2009).
Preitner, N. et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002).
Raghuram, S. et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol. 14, 1207–1213 (2007).
Chomez, P. et al. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(α) orphan receptor. Development 127, 1489–1498 (2000).
Harding, H.P. & Lazar, M.A. The orphan receptor Rev-ErbA α activates transcription via a novel response element. Mol. Cell. Biol. 13, 3113–3121 (1993).
Adelmant, G., Begue, A., Stehelin, D. & Laudet, V. A functional Rev-erb α responsive element located in the human Rev-erb α promoter mediates a repressing activity. Proc. Natl. Acad. Sci. USA 93, 3553–3558 (1996).
Yin, L., Wang, J., Klein, P.S. & Lazar, M.A. Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311, 1002–1005 (2006).
Flavell, S.W. & Greenberg, M.E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 31, 563–590 (2008).
Thompson, K.R. et al. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron 44, 997–1009 (2004).
An, J.J. et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175–187 (2008).
Ferkany, J.W., Zaczek, R. & Coyle, J.T. The mechanism of kainic acid neurotoxicity. Nature 308, 561–562 (1984).
Zaczek, R., Nelson, M. & Coyle, J.T. Kainic acid neurotoxicity and seizures. Neuropharmacology 20, 183–189 (1981).
Pardee, K.I. et al. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. PLoS Biol. 7, e43 (2009).
Ueda, H.R. et al. A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 (2002).
Lamont, E.W., Robinson, B., Stewart, J. & Amir, S. The central and basolateral nuclei of the amygdala exhibit opposite diurnal rhythms of expression of the clock protein Period2. Proc. Natl. Acad. Sci. USA 102, 4180–4184 (2005).
Wakamatsu, H. et al. Restricted feeding–induced anticipatory activity rhythm is associated with a phase-shift of the expression of mPer1 and mPer2 mRNA in the cerebral cortex and hippocampus but not in the suprachiasmatic nucleus of mice. Eur. J. Neurosci. 13, 1190–1196 (2001).
Chaudhury, D., Loh, D.H., Dragich, J.M., Hagopian, A. & Colwell, C.S. Select cognitive deficits in vasoactive intestinal peptide deficient mice. BMC Neurosci. 9, 63 (2008).
Abraham, U., Prior, J.L., Granados-Fuentes, D., Piwnica-Worms, D.R. & Herzog, E.D. Independent circadian oscillations of Period1 in specific brain areas in vivo and in vitro. J. Neurosci. 25, 8620–8626 (2005).
Mendoza, J., Pevet, P., Felder-Schmittbuhl, M.P., Bailly, Y. & Challet, E. The cerebellum harbors a circadian oscillator involved in food anticipation. J. Neurosci. 30, 1894–1904 (2010).
Abe, H., Honma, S., Namihira, M., Masubuchi, S. & Honma, K. Behavioral rhythm splitting in the CS mouse is related to clock gene expression outside the suprachiasmatic nucleus. Eur. J. Neurosci. 14, 1121–1128 (2001).
Gilestro, G.F., Tononi, G. & Cirelli, C. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324, 109–112 (2009).
Hastings, M.H., Reddy, A.B. & Maywood, E.S. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661 (2003).
Triqueneaux, G. et al. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 33, 585–608 (2004).
Nadif Kasri, N., Nakano-Kobayashi, A., Malinow, R., Li, B. & Van Aelst, L. The Rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev. 23, 1289–1302 (2009).
Vyazovskiy, V.V., Cirelli, C., Pfister-Genskow, M., Faraguna, U. & Tononi, G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat. Neurosci. 11, 200–208 (2008).
Donlea, J.M., Ramanan, N. & Shaw, P.J. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108 (2009).
Flora, A. et al. Sp proteins and Phox2b regulate the expression of the human Phox2a gene. J. Neurosci. 21, 7037–7045 (2001).
Battaglioli, E. et al. Expression and transcriptional regulation of the human alpha3 neuronal nicotinic receptor subunit in T lymphocyte cell lines. J. Neurochem. 71, 1261–1270 (1998).
Sala, C., Rudolph-Correia, S. & Sheng, M. Developmentally regulated NMDA receptor–dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J. Neurosci. 20, 3529–3536 (2000).
Passafaro, M., Nakagawa, T., Sala, C. & Sheng, M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature 424, 677–681 (2003).
Saglietti, L. et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 54, 461–477 (2007).
Acknowledgements
The authors thank D. Ward for assisting with the manuscript. The authors thank E. Challet, D. Ciocca and S. Reibel-Foisset for advice with the circadian experiments. M.P. was supported by Telethon Italy (S01014TELU), Fondazione Cariplo (2008-2318), Fondazione Mariani, Project TerDisMental, ID 16983 - Rif. SAL-50. C.S. was supported by Telethon-Italy grant GGP09196, Fondazione CARIPLO project no. 2009.264, Ricerca Scientifica a Tema Libero (RSTL) Consiglio Nazionale delle Ricerche (CNR), Regione Lombardia Project no. SAL-50-16983 TERDISMENTAL and an Italian Institute of Technology Seed Grant. P.B., O.D. and J.C. are supported by the French National Research Agency (ANR-06-Neuro-003-02, ANR-08-MNPS-037-04), European Union (Gencodys, FP7 241995), Fondation Jérôme Lejeune and INSERM. C.L. was supported by the “Fondation pour La Recherche Médicale”.
Author information
Authors and Affiliations
Contributions
P.V. conducted all the experiments in COS7 cells, in hippocampal neurons and in vivo. M.K., O.D. and C.L. conducted the experiments on circadian cycle. S.B. and A.G. prepared mutants for yeast two-hybrid screening. R.B. supervised the experiments with luciferase assays. J.C., P.B. and C.S. supervised the project. M.P. wrote the manuscript and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 4472 kb)
Rights and permissions
About this article
Cite this article
Valnegri, P., Khelfaoui, M., Dorseuil, O. et al. A circadian clock in hippocampus is regulated by interaction between oligophrenin-1 and Rev-erbα. Nat Neurosci 14, 1293–1301 (2011). https://doi.org/10.1038/nn.2911
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2911
This article is cited by
-
Exome sequencing identifies novel and known mutations in families with intellectual disability
BMC Medical Genomics (2021)
-
Role of Per3, a circadian clock gene, in embryonic development of mouse cerebral cortex
Scientific Reports (2019)
-
Circadian blueprint of metabolic pathways in the brain
Nature Reviews Neuroscience (2019)
-
Role of a circadian-relevant gene NR1D1 in brain development: possible involvement in the pathophysiology of autism spectrum disorders
Scientific Reports (2017)
-
Long days enhance recognition memory and increase insulin-like growth factor 2 in the hippocampus
Scientific Reports (2017)