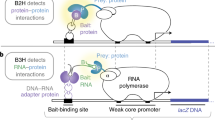
Abstract
Bacterial-based interaction trap systems provide a powerful method to identify interacting macromolecules. When carried out in the context of a genetic selection, interacting pairs can be rapidly isolated from large combinatorial libraries. This technology has been adapted to allow the identification of DNA-binding sequences for a transcription factor (TF) from a large randomized library. This procedure uses a library of randomized binding sites upstream of a cocistronic HIS3-URA3 reporter cassette. The URA3 reporter allows self-activating sequences to be removed from the library through counter-selection. The HIS3 reporter allows sequences that are recognized by a TF to be isolated from the library, where transcriptional activation is mediated by fusion of the TF to the α-subunit of RNA polymerase. This technology can be used to characterize monomeric, homodimeric and heterodimeric DNA-binding domains and, once a suitable library is constructed, binding sites can be identified in approximately 10 d. The bacterial one-hybrid system allows larger libraries to be searched than the corresponding yeast one-hybrid system and, unlike SELEX, it does not require purification of the TF(s). The complexity of the binding site libraries that can be searched using the bacterial system is, however, more limited than SELEX, and some eukaryotic factors may not express or fold efficiently in the bacterial system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Joung, J.K., Ramm, E.I. & Pabo, C.O. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl. Acad. Sci. USA 97, 7382–7387 (2000).
Meng, X., Smith, R.M., Giesecke, A.G., Joung, J.K. & Wolfe, S.A. Counter-selectable marker for bacterial-based interaction trap systems. Biotechniques 40, 179–184 (2006).
Nickels, B.E. et al. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl. Acad. Sci. USA 102, 4488–4493 (2005).
Vallet-Gely, I., Donovan, K.E., Fang, R., Joung, J.K. & Dove, S.L. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 102, 11082–11087 (2005).
Giesecke, A., Fang, R. & Joung, J. Synthetic protein–protein interaction domains created by shuffling Cys2His2 zinc-fingers. Mol. Syst. Biol. 2, published online 31 March 2006 (doi:10.1038/msb4100053).
Hurt, J.A., Thibodeau, S.A., Hirsh, A.S., Pabo, C.O. & Joung, J.K. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc. Natl. Acad. Sci. USA 100, 12271–12276 (2003).
Meng, X., Brodsky, M.H. & Wolfe, S.A. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23, 988–994 (2005).
Durai, S., Bosley, A., Abulencia, A.B., Chandrasegaran, S. & Ostermeier, M. A bacterial one-hybrid selection system for interrogating zinc finger-DNA interactions. Comb. Chem. High Throughput Screen 9, 301–311 (2006).
Althoff, E.A. & Cornish, V.W. A bacterial small-molecule three-hybrid system. Angew Chem. Int. Ed. Engl. 41, 2327–2330 (2002).
Dove, S.L. & Hochschild, A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 12, 745–754 (1998).
Dove, S.L., Joung, J.K. & Hochschild, A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386, 627–630 (1997).
Wilson, T.E., Fahrner, T.J., Johnston, M. & Milbrandt, J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252, 1296–1300 (1991).
Deplancke, B., Dupuy, D., Vidal, M. & Walhout, A.J. A gateway-compatible yeast one-hybrid system. Genome Res. 14, 2093–2101 (2004).
Wright, W.E. & Funk, W.D. CASTing for multicomponent DNA-binding complexes. Trends Biochem. Sci. 18, 77–80 (1993).
Roulet, E. et al. High-throughput SELEX-SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 20, 831–835 (2002).
Hu, J.C., Kornacker, M.G. & Hochschild, A. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods 20, 80–94 (2000).
Serebriiskii, I.G. et al. A combined yeast/bacteria two-hybrid system: development and evaluation. Mol. Cell. Proteomics 4, 819–826 (2005).
Jen-Jacobson, L. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers 44, 153–180 (1997).
Thibodeau, S.A., Fang, R. & Joung, J.K. High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. Biotechniques 36, 410–415 (2004).
Solomon, D.L., Amati, B. & Land, H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 21, 5372–5376 (1993).
Yan, R., Small, S., Desplan, C., Dearolf, C.R. & Darnell, J.E. Jr. Identification of a Stat gene that functions in Drosophila development. Cell 84, 421–430 (1996).
Patrick, W.M., Firth, A.E. & Blackburn, J.M. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. 16, 451–457 (2003).
Bosley, A.D. & Ostermeier, M. Mathematical expressions useful in the construction, description and evaluation of protein libraries. Biomol. Eng. 22, 57–61 (2005).
Bailey, T.L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Liu, X., Brutlag, D.L. & Liu, J.S. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 6, 127–138 (2001).
Bailey, T.L. & Elkan, C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3, 21–29 (1995).
Tompa, M. et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 23, 137–144 (2005).
Hu, J., Li, B. & Kihara, D. Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 33, 4899–4913 (2005).
Schneider, T.D. & Stephens, R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Acknowledgements
We would like to thank J. McNulty and M. Noyes for critiquing this manuscript. This work was supported by a National Institutes of Health grant 1R01GM068110.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have a pending patent application on related subject matter (USPTO).
Supplementary information
Supplementary Figure 1
Self-activating sequences isolated from the binding site selections. (PDF 3712 kb)
Rights and permissions
About this article
Cite this article
Meng, X., Wolfe, S. Identifying DNA sequences recognized by a transcription factor using a bacterial one-hybrid system. Nat Protoc 1, 30–45 (2006). https://doi.org/10.1038/nprot.2006.6
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.6
This article is cited by
-
Engineered dual selection for directed evolution of SpCas9 PAM specificity
Nature Communications (2021)
-
DNA–protein interaction: identification, prediction and data analysis
Molecular Biology Reports (2019)
-
A high throughput approach for the generation of orthogonally interacting protein pairs
Scientific Reports (2018)
-
Evolved Cas9 variants with broad PAM compatibility and high DNA specificity
Nature (2018)
-
De-novo protein function prediction using DNA binding and RNA binding proteins as a test case
Nature Communications (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.