Abstract
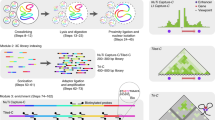
The pioneering chromosome conformation capture (3C) method provides the opportunity to study chromosomal folding in the nucleus. It is based on formaldehyde cross-linking of living cells followed by enzyme digestion, intramolecular ligation and quantitative (Q)-PCR analysis. However, 3C requires prior knowledge of the bait and interacting sequence (termed interactor) rendering it less useful for genome-wide studies. As several recent reports document, this limitation has been overcome by exploiting a circular intermediate in a variant of the 3C method, termed 4C (for circular 3C). The strategic positioning of primers within the bait enables the identification of unknown interacting sequences, which form part of the circular DNA. Here, we describe a protocol for our 4C method, which produces a high-resolution interaction map potentially suitable for the analysis of cis-regulatory elements and for comparison with chromatin marks obtained by chromatin immunoprecipitation (ChIP) on chip at the sites of interaction. Following optimization of enzyme digestions and amplification conditions, the protocol can be completed in 2–3 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Branco, M.R. & Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4, e138 (2006).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Dekker, J. The three 'C' s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3, 17–21 (2006).
Hagege, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Wurtele, H. & Chartrand, P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 14, 477–495 (2006).
Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 (2006).
Ohlsson, R. & Göndör, A. The 4C technique: The 'Rosetta stone' for genome biology in 3D? Curr. Opin. Cell Biol. 19, 321–325 (2007).
Ling, J.Q. et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 (2006).
Kurukuti, S. et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 103, 10684–10689 (2006).
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Kiernan, J.A. Formaldehyde, formalin, paraformaldehyde and glutaraldehyde: what they are and what they do. Microscopy Today 8, 8–12 (2000).
Acknowledgements
We gratefully acknowledge help with the processing of NimbleGen data from Kuljeet Singh and discussions with many members of the RO lab. This work was supported by grants from HEROIC (EU), Swedish Research Council, Swedish Cancer Foundation, Swedish Pediatric Cancer Foundation and Lundberg's Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Göndör, A., Rougier, C. & Ohlsson, R. High-resolution circular chromosome conformation capture assay. Nat Protoc 3, 303–313 (2008). https://doi.org/10.1038/nprot.2007.540
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.540
This article is cited by
-
The Nodewalk assay to quantitate chromatin fiber interactomes in very small cell populations
Nature Protocols (2023)
-
WNT signaling and AHCTF1 promote oncogenic MYC expression through super-enhancer-mediated gene gating
Nature Genetics (2019)
-
Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2
BMC Cancer (2016)
-
Distal regulation of c-myb expression during IL-6-induced differentiation in murine myeloid progenitor M1 cells
Cell Death & Disease (2016)
-
Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia
Leukemia (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.