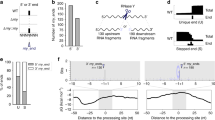
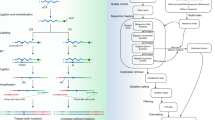
Abstract
We have developed a novel approach called parallel analysis of RNA ends (PARE) for high-throughput identification of microRNA (miRNA) targets and diverse applications for the study of the RNA degradome. The method described here comprises a modified 5′-rapid amplification of cDNA ends, deep sequencing of 3′ cleavage products of mRNA and bioinformatic analysis. Following RNA extraction and isolation of polyadenylated RNA, a 5′-RNA adapter that includes an MmeI recognition site is ligated to 5′-monophosphorylated products, which contain mRNA fragments generated through miRNA-induced cleavage. The ligated products are reverse-transcribed, slightly amplified and cleaved with MmeI. The 5′ equally-sized fragments are gel-selected, ligated to a 3′ double-stranded DNA adapter and PCR-amplified. Following gel purification, the products are subjected to deep sequencing. The data are then matched to cDNAs and analyzed through bioinformatics filters. We describe the high-throughput protocol in detail and indicate alternative uses for PARE. The procedure presented here can be accomplished in 6–7 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Palatnik, J.F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263 (2003).
Llave, C., Xie, Z., Kasschau, K.D. & Carrington, J.C. Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 (2002).
Brodersen, P. et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 (2008).
Aukerman, M.J. & Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741 (2003).
Rhoades, M.W. et al. Prediction of plant microRNA targets. Cell 110, 513–520 (2002).
Bonnet, E., Wuyts, J., Rouze, P. & Van de Peer, Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101, 11511–11516 (2004).
Zhang, Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 33, W701–704 (2005).
Jones-Rhoades, M.W. & Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799 (2004).
Schwab, R. et al. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527 (2005).
Jones-Rhoades, M.W., Bartel, D.P. & Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53 (2006).
Ebert, M.S., Neilson, J.R. & Sharp, P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 (2007).
Elbashir, S.M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001).
Gregory, B.D. et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14, 854–866 (2008).
German, M.A. et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946 (2008).
Addo-Quaye, C., Eshoo, T.W., Bartel, D.P. & Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762 (2008).
Sambrook, J., Fritsch, D.F. & Maniatis, T. Molecular Cloning: A Laboratory Manual 2nd edn. (Cold Spring Harbor Press, Cold Spring Harbor, NY, 1989).
Yekta, S., Shih, I.H. & Bartel, D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594–596 (2004).
Acknowledgements
We are grateful to Shawn Thatcher for help in editing the manuscript. This work was supported primarily by DOE no. DE-FG02-04ER15541 and NSF no. 0445638 (P.J.G.), with additional support from USDA no. 2007-01991 (P.J.G.), NSF no. 0548569 (P.J.G and B.C.M.), NSF no. 0321437 (B.C.M.) and NIH P20 RR16472-04.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
S.L and G.S. are employees of Illumina, which is the company that has commercialized the SBS sequencing technology. Our paper describes the use of this technology for the analysis of miRNA-target RNA pairs and the RNA degradome we demonstrated this approach in Arabidopsis, but the method has broad applicability.
Rights and permissions
About this article
Cite this article
German, M., Luo, S., Schroth, G. et al. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4, 356–362 (2009). https://doi.org/10.1038/nprot.2009.8
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.8
This article is cited by
-
Heat-responsive microRNAs participate in regulating the pollen fertility stability of CMS-D2 restorer line under high-temperature stress
Biological Research (2023)
-
Cut site preference allows influenza A virus PA-X to discriminate between host and viral mRNAs
Nature Microbiology (2023)
-
MicroRNA miR394 regulates flowering time in Arabidopsis thaliana
Plant Cell Reports (2022)
-
Degradome sequencing-based identification of phasiRNAs biogenesis pathways in Oryza sativa
BMC Genomics (2021)
-
Pi-starvation induced transcriptional changes in barley revealed by a comprehensive RNA-Seq and degradome analyses
BMC Genomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.