Key Points
-
Ribosome biogenesis and translation are regulated at multiple levels and are associated with accurate cell growth and proliferation. The loss of key checkpoints during protein synthesis might contribute to the initiation and progression of cancer.
-
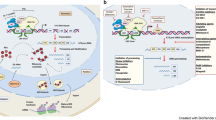
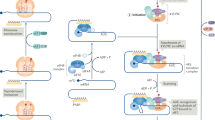
During specific phases of the cell cycle, the synthesis of rRNA, as well as components of the protein synthesis machinery, is initiated by the phosphorylation of key transcription factors that regulate polymerase I (Pol I) and Pol III activity, respectively. This tight link between cell-cycle progression and protein synthesis exists to ensure accurate cell growth and proliferation, which might be lost in cancer cells.
-
p53 and retinoblastoma (RB) repress Pol I and Pol III transcription. In cancer cells, which harbour inactivating mutations in these tumour suppressors, deregulation of Pol I and Pol III activity might contribute to tumorigenesis.
-
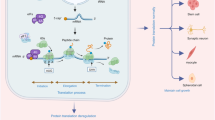
Several ribosomal proteins are overexpressed in a variety of tumours. It remains to be determined whether this represents a cause or consequence of tumour formation. Increased phosphorylation of the S6 ribosomal protein is thought to result in enhanced translation of specific mRNAs. This raises the possibility that deregulation of ribosomal proteins in tumours might affect the translation of specific target mRNAs.
-
MYC and PTEN act as master regulators of ribosome biogenesis and translation control. Their deregulation in tumour cells increases the expression and activity of components of the translation apparatus. It remains to be determined which of the downstream targets of MYC and PTEN involved in controlling protein synthesis are directly responsible for tumour susceptibility.
-
Further investigation will be needed to clarify to what extent deregulation in total or specific translation of mRNAs contributes to tumorigenesis. Although mutations in genes that are directly responsible for ribosome biogenesis, such as those encoding the ribosomal protein S19 and DKC1 (the enzyme that modifies rRNA), have been found in cancer susceptibility syndromes, the molecular mechanisms by which these proteins cause cancer remain largely unknown.
-
Components of the translation machinery that are overexpressed or deregulated in cancer cells could represent targets for cancer therapy. The macrolide rapamycin, which affects the translation machinery, has already been used in clinical trials as a tumour inhibitory agent.
Abstract
Ribosome biogenesis and translation control are essential cellular processes that are governed at numerous levels. Several tumour suppressors and proto-oncogenes have been found either to affect the formation of the mature ribosome or to regulate the activity of proteins known as translation factors. Disruption in one or more of the steps that control protein biosynthesis has been associated with alterations in the cell cycle and regulation of cell growth. Therefore, certain tumour suppressors and proto-oncogenes might regulate malignant progression by altering the protein synthesis machinery. Although many studies have correlated deregulation of protein biosynthesis with cancer, it remains to be established whether this translates directly into an increase in cancer susceptibility, and under what circumstances.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Gani, R. The nucleoli of cultured human lymphocytes. I. Nucleolar morphology in relation to transformation and the DNA cycle. Exp. Cell Res. 97, 249–258 (1976).
Heiss, N. S. et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genet. 19, 32–38 (1998).
Ruggero, D. et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299, 259–262 (2003). In this study, animal models for dyskeratosis congenita were generated, which are hypomorphic mutants for the pseudouridine synthase, DKC1. These mice are faithful models of the disease and show marked cancer susceptibility associated with an impairment in ribosome biogenesis.
Draptchinskaia, N. et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature Genet. 21, 169–175 (1999). Mutations in the ribosomal protein S19 were found to be associated with a disease characterized by an increased susceptibility to cancer.
Pardee, A. B. G1 events and regulation of cell proliferation. Science 246, 603–608 (1989).
Pyronnet, S. & Sonenberg, N. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 11, 13–18 (2001).
Grummt, I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62, 109–154 (1999).
Grummt, I., Smith, V. A. & Grummt, F. Amino acid starvation affects the initiation frequency of nucleolar RNA polymerase. Cell 7, 439–445 (1976).
Kief, D. R. & Warner, J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 1, 1007–1015 (1981).
Klein, J. & Grummt, I. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl Acad. Sci. USA 96, 6096–6101 (1999). An important paper showing that UBF activity is regulated in a cell-cycle-dependent manner and that fluctuations in rRNA synthesis during the cell cycle are controlled by UBF phosphorylation.
Cavanaugh, A. H. et al. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 9, 177–180 (1995). A landmark paper which shows that the tumour-suppressor gene Rb directly represses transcription of rRNA through inhibition of UBF function. There is an accumulation of Rb in the nucleolus of differentiated cells, indicating that Rb might exert some of its growth-suppressive properties by regulating rRNA synthesis.
Beckmann, H., Chen, J. L., O'Brien, T. & Tjian, R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science 270, 1506–1509 (1995).
Brandenburger, Y., Jenkins, A., Autelitano, D. J. & Hannan, R. D. Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J. 15, 2051–2053 (2001).
Bell, S. P., Learned, R. M., Jantzen, H. M. & Tjian, R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241, 1192–1197 (1988).
antzen, H. M., Admon, A., Bell, S. P. & Tjian, R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344, 830–836 (1990).
Read, C. et al. High resolution studies of the Xenopus laevis ribosomal gene promoter in vivo and in vitro. J. Biol. Chem. 267, 10961–10967 (1992).
Voit, R., Hoffmann, M. & Grummt, I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18, 1891–1899 (1999).
Kihm, A. J., Hershey, J. C., Haystead, T. A., Madsen, C. S. & Owens, G. K. Phosphorylation of the rRNA transcription factor upstream binding factor promotes its association with TATA binding protein. Proc. Natl Acad. Sci. USA 95, 14816–14820 (1998).
Tuan, J. C., Zhai, W. & Comai, L. Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 19, 2872–2879 (1999).
Voit, R. & Grummt, I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl Acad. Sci. USA 98, 13631–13636 (2001).
Stefanovsky, V. Y. et al. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8, 1063–1073 (2001). The study shows that the activation of the ERK1/2 MAP kinases by growth factors leads to rRNA transcription through a mechanism that is dependent on UBF phosphorylation. This study provides a link between growth-factor signalling and increased ribosome production.
O'Mahony, D. J., Xie, W. Q., Smith, S. D., Singer, H. A. & Rothblum, L. I. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J. Biol. Chem. 267, 35–38 (1992).
Voit, R. et al. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 11, 2211–2218 (1992).
Voit, R., Kuhn, A., Sander, E. E. & Grummt, I. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23, 2593–2599 (1995).
Wang, T. C. et al. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369, 669–671 (1994).
Bortner, D. M. & Rosenberg, M. P. Induction of mammary gland hyperplasia and carcinomas in transgenic mice expressing human cyclin E. Mol. Cell. Biol. 17, 453–459 (1997).
Navolanic, P. M., Steelman, L. S. & McCubrey, J. A. EGFR family signaling and its association with breast cancer development and resistance to chemotherapy. Int. J. Oncol. 22, 237–252 (2003).
Cavanaugh, A. H. et al. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277, 27423–27432 (2002).
Yuan, X., Zhao, J., Zentgraf, H., Hoffmann-Rohrer, U. & Grummt, I. Multiple interactions between RNA polymerase I, TIF-IA and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3, 1082–1087 (2002).
Classon, M. & Harlow, E. The retinoblastoma tumour suppressor in development and cancer. Nature Rev. Cancer 2, 910–917 (2002).
Voit, R., Schafer, K. & Grummt, I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell. Biol. 17, 4230–4237 (1997).
Hannan, K. M. et al. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 19, 4988–4999 (2000).
Ciarmatori, S. et al. Overlapping functions of the pRb family in the regulation of rRNA synthesis. Mol. Cell. Biol. 21, 5806–5814 (2001).
Hannan, K. M. et al. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene 19, 3487–3497 (2000).
Zhai, W. & Comai, L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20, 5930–5938 (2000). Shows that the tumour suppressor p53 can directly regulate rRNA synthesis through its interaction with UBF.
Cordon-Cardo, C. et al. Cooperative effects of p53 and pRB alterations in primary superficial bladder tumors. Cancer Res. 57, 1217–1221 (1997).
White, R. J., Trouche, D., Martin, K., Jackson, S. P. & Kouzarides, T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature 382, 88–90 (1996).
Sutcliffe, J. E., Brown, T. R., Allison, S. J., Scott, P. H. & White, R. J. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 20, 9192–9202 (2000).
Cairns, C. A. & White, R. J. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 17, 3112–3123 (1998).
Kaye, F. J., Kratzke, R. A., Gerster, J. L. & Horowitz, J. M. A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc. Natl Acad. Sci. USA 87, 6922–6926 (1990).
Larminie, C. G., Alzuherri, H. M., Cairns, C. A., McLees, A. & White, R. J. Transcription by RNA polymerases I and III: a potential link between cell growth, protein synthesis and the retinoblastoma protein. J. Mol. Med. 76, 94–103 (1998).
Johnson, L. F., Williams, J. G., Abelson, H. T., Green, H. & Penman, S. Changes in RNA in relation to growth of the fibroblast. III. Posttranscriptional regulation of mRNA formation in resting and growing cells. Cell 4, 69–75 (1975).
Baxter, G. C. & Stanners, C. P. The effect of protein degradation on cellular growth characteristics. J. Cell. Physiol. 96, 139–145 (1978).
Frank, D. J., Edgar, B. A. & Roth, M. B. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129, 399–407 (2002).
Frank, D. J. & Roth, M. B. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J. Cell Biol. 140, 1321–1329 (1998).
Fatica, A. & Tollervey, D. Making ribosomes. Curr. Opin. Cell. Biol. 14, 313–318 (2002).
Draper, D. E. & Reynaldo, L. P. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 27, 381–388 (1999).
Maguire, B. A. & Zimmermann, R. A. The ribosome in focus. Cell 104, 813–816 (2001).
Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002).
Ferrari, S. et al. Noncoordinated expression of S6, S11, and S14 ribosomal protein genes in leukemic blast cells. Cancer Res. 50, 5825–5828 (1990).
Loging, W. T. & Reisman, D. Elevated expression of ribosomal protein genes L37, RPP-1, and S2 in the presence of mutant p53. Cancer Epidemiol. Biomarkers Prev. 8, 1011–1016 (1999).
Kondoh, N. et al. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res. 21, 2429–2433 (2001).
Zhang, L. et al. Gene expression profiles in normal and cancer cells. Science 276, 1268–1272 (1997).
Alon, U. et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl Acad. Sci. USA 96, 6745–6750 (1999).
Uechi, T., Tanaka, T. & Kenmochi, N. A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics 72, 223–230 (2001).
Bassoe, C. F., Bruserud, O., Pryme, I. F. & Vedeler, A. Ribosomal proteins sustain morphology, function and phenotype in acute myeloid leukemia blasts. Leuk. Res. 22, 329–339 (1998).
Ferrari, S. et al. Abundance of the primary transcript and its processed product of growth-related genes in normal and leukemic cells during proliferation and differentiation. Cancer Res. 52, 11–16 (1992).
Warner, J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999).
Rotenberg, M. O., Moritz, M. & Woolford, J. L. Jr. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2, 160–172 (1988).
Kongsuwan, K. et al. A Drosophila Minute gene encodes a ribosomal protein. Nature 317, 555–558 (1985).
Kay, M. A. & Jacobs-Lorena, M. Developmental genetics of ribosome synthesis in Drosophila. Trends Genet. 3, 347–351 (1987).
Volarevic, S. et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288, 2045–2047 (2000).
Terada, N. et al. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl Acad. Sci. USA 91, 11477–11481 (1994).
Jefferies, H. B., Reinhard, C., Kozma, S. C. & Thomas, G. Rapamycin selectively represses translation of the 'polypyrimidine tract' mRNA family. Proc. Natl Acad. Sci. USA 91, 4441–4445 (1994).
Jefferies, H. B. et al. Rapamycin suppresses 5′ TOP mRNA translation through inhibition of p70s6k. EMBO J. 16, 3693–3704 (1997).
Meyuhas, O. & Hornstein, E. in Translational Control of Gene Expression (eds Sonenberg, N., Hershey, J. W. B. & Mathews, M. B.) (Cold Spring Harbor Laboratory Press, New York, 2000).
Anand, N. et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nature Genet. 31, 301–305 (2002).
Thomas, G., Siegmann, M. & Gordon, J. Multiple phosphorylation of ribosomal protein S6 during transition of quiescent 3T3 cells into early G1, and cellular compartmentalization of the phosphate donor. Proc. Natl Acad. Sci. USA 76, 3952–3956 (1979).
Gressner, A. M. & Wool, I. G. The phosphorylation of liver ribosomal proteins in vivo. Evidence that only a single small subunit protein (S6) is phosphorylated. J. Biol. Chem. 249, 6917–6925 (1974).
Kozma, S. C. Thomas G. p70s6k/p85s6k: mechanism of activation and role in mitogenesis. Cancer Biol. 5, 255–260 (1994).
Montagne, J. et al. Drosophila S6 kinase: a regulator of cell size. Science 285, 2126–2129 (1999).
Shima, H. et al. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17, 6649–6659 (1998).
Kawasome, H. et al. Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity. Proc. Natl Acad. Sci. USA 95, 5033–5038 (1998).
Stolovich, M. et al. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22, 8101–8113 (2002).
Naora, H., Takai, I., Adachi, M. & Naora, H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J. Cell. Biol. 141, 741–753 (1998).
Kim, J. et al. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J. Biol. Chem. 270, 13620–13629 (1995).
Boxer, L. M. & Dang, C. V. Translocations involving c-myc and c-myc function. Oncogene 20, 5595–5610 (2001).
Coller, H. A. et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl Acad. Sci. USA 97, 3260–3265 (2000).
Boon, K. et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 20, 1383–1393 (2001). SAGE analysis of MYCN target genes in human neuroblastoma cells reveals that most MYC target genes are involved in ribosome biogenesis and protein synthesis.
Menssen, A. & Hermeking, H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl Acad. Sci. USA 99, 6274–6279 (2002).
Iritani, B. M. & Eisenman, R. N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl Acad. Sci. USA 96, 13180–13185 (1999). The direct overexpression of c-Myc during B-cell development results in increased cell growth that is independent of the cell cycle and correlates with an increase in total protein synthesis in the cell.
Kim, S., Li, Q., Dang, C. V. & Lee, L. A. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl Acad. Sci. USA 97, 11198–11202 (2000).
Iritani, B. M. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 21, 4820–4830 (2002).
Piedra, M. E., Delgado, M. D., Ros, M. A. & Leon, J. c-Myc overexpression increases cell size and impairs cartilage differentiation during chick limb development. Cell Growth Differ. 13, 185–193 (2002).
Schmidt, E. V. The role of c-myc in cellular growth control. Oncogene 18, 2988–2996 (1999).
Kozma, S. C. & Thomas, G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24, 65–71 (2002).
Di Cristofano, A., Pesce, B., Cordon-Cardo, C. & Pandolfi, P. P. Pten is essential for embryonic development and tumour suppression. Nature Genet. 19, 348–355 (1998).
Di Cristofano, A., De Acetis, M., Koff, A., Cordon-Cardo, C. & Pandolfi, P. P. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nature Genet. 27, 222–224 (2001).
Vogt, P. K. PI 3-kinase, mTOR, protein synthesis and cancer. Trends Mol. Med. 7, 482–484 (2001).
Backman, S., Stambolic, V. & Mak, T. PTEN function in mammalian cell size regulation. Curr. Opin. Neurobiol. 12, 516–522 (2002).
Maehama, T. & Dixon, J. E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378 (1998).
Myers, M. P. et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl Acad. Sci. USA. 95, 13513–13518 (1998).
Brown, E. J. et al. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377, 441–446 (1995).
Burnett, P. E., Barrow, R. K., Cohen, N. A., Snyder, S. H. & Sabatini, D. M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl Acad. Sci. USA 95, 1432–1437 (1998).
Isotani, S. et al. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase α in vitro. J. Biol. Chem. 274, 34493–34498 (1999).
Sabatini, D. M. Interaction of RAFT1 with gephyrin required for rapamycin-sensitive signaling. Science 284, 1161–1164 (1999).
Neshat, M. S. et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl Acad. Sci. USA 98, 10314–10319 (2001).
Podsypanina, K. et al. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proc. Natl Acad. Sci. USA 98, 10320–10325 (2001). References 97 and 98 report that Pten-deficient tumours show an increase in mTOR activation and an upregulation in p70S6k activity. These tumours can be effectively treated with the macrolide rapamycin, the cellular target of which is mTOR.
Gingras, A. C. et al. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 12, 502–513 (1998).
Sekulic, A. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 60, 3504–3513 (2000).
Pullen, N. et al. Phosphorylation and activation of p70s6k by PDK1. Science 279, 707–710 (1998).
Aoki, M., Blazek, E. & Vogt, P. K. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc. Natl Acad. Sci. USA 98, 136–141 (2001).
Podsypanina, K. et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl Acad. Sci. USA 96, 1563–1568 (1999).
Suzuki, A. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 8, 1169–1178 (1998).
Gomez, M., Sampson, J. & Whittemore, V. The Tuberous Sclerosis Complex (Oxford Univ. Press, 1999).
Kwiatkowski, D. J. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11, 525–534 (2002).
Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature Cell Biol. 4, 648–657 (2002).
Gao, X. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nature Cell Biol. 4, 699–704 (2002).
Tee, A. R. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl Acad. Sci. USA 99, 13571–13576 (2002).
Radimerski, T., Montagne, J., Hemmings-Mieszczak, M. & Thomas, G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 16, 2627–2632 (2002).
Goncharova, E. A. et al. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM). J. Biol. Chem. 277, 30958–30967 (2002).
Jaeschke, A. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 159, 217–224 (2002).
Lazaris-Karatzas, A., Montine, K. S. & Sonenberg, N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345, 544–547 (1990). The first demonstration that a translation initiation factor shows oncogenic properties when constitutively overexpressed.
De Benedetti, A. & Harris, A. L. eIF4E expression in tumors: its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 31, 59–72 (1999).
Gingras, A. C., Raught, B. & Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 (1999).
Ni, J., Tien, A. L. & Fournier, M. J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573 (1997).
Lafontaine, D. L., Bousquet-Antonelli, C., Henry, Y., Caizergues-Ferrer, M. & Tollervey, D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12, 527–537 (1998).
Dokal, I. Dyskeratosis congenita in all its forms. Br. J. Haematol. 110, 768–779 (2000).
Giordano, E., Peluso, I., Senger, S. & Furia, M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 144, 1123–1133 (1999).
Zebarjadian, Y., King, T., Fournier, M. J., Clarke, L. & Carbon, J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19, 7461–7472 (1999).
Mitchell, J. R., Wood, E. & Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999).
Rudolph, K. L. et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712 (1999).
Decatur, W. A. & Fournier, M. J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002).
Kullmann, M., Gopfert, U., Siewe, B. & Hengst, L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev. 16, 3087–3099 (2002).
Cornelis, S. et al. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5, 597–605 (2000).
Hoang, C. & Ferre-D'Amare, A. R. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107, 929–939 (2001).
Vagner, S., Galy, B. & Pyronnet, S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2, 893–898 (2001).
Da Costa, L., Willig, T. N., Fixler, J., Mohandas, N. & Tchernia, G. Diamond-Blackfan anemia. Curr. Opin. Pediatr. 13, 10–15 (2001).
Willis, A. E. Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell. Biol. 31, 73–86 (1999).
Spence, J. et al. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102, 67–76 (2000).
Strezoska, Z., Pestov, D. G. & Lau, L. F. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20, 5516–5528 (2000).
Pestov, D. G., Strezoska, Z. & Lau, L. F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 21, 4246–4255 (2001). A defect in ribosome rRNA processing can directly affect cell-cycle progression through a p53-dependent process. This is a hallmark study proposing that a nucleolar stress affects cellular proliferation through crosstalk between the ribosome and the cell cycle.
Hidalgo, M. & Rowinsky, E. K. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 19, 6680–6686 (2000).
Decatur, W. A. & Fournier, M. J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278, 695–698 (2003).
Manning, B. D., Tee, A. R., Logsdon, M. N., Blenis, J. & Cantley, L. C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 (2002).
Potter, C. J., Pedraza, L. G. & Xu, T. Akt regulates growth by directly phosphorylating Tsc2. Nature Cell Biol. 4, 658–665 (2002). References 135 and 136 show that AKT phosphorylates TSC2 and thereby inhibits its function as negative regulator of p70S6k.
Acknowledgements
We are indebted to M. Barna for input, discussion and critical reading of the review. We apologize to the many scientists whose work we did not cite due to space constraints. This work is supported by the National Cancer Institute, the Mouse Model of Human Cancer Consortium (MMHCC) and the Leukemia Lymphoma Society (LLS).
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
CancerNet
LocusLink
OMIM
Glossary
- NUCLEOLUS
-
The nucleolus is a suborganelle of the nucleus where rRNA is transcribed, processed and assembled into ribosomal subumits. Ribosomal proteins are added to rRNAs within the nucleolus.
- CRYO-ELECTRON MICROSCOPY
-
A technique that is used to study the three-dimensional structures of biomolecules. A sample is rapidly frozen in liquid helium to preserve its structure and then examined in the frozen, hydrated state in a cryoelectron microscope.
- RAPAMYCIN
-
Rapamycin is a macrolide with immunosuppressant properties and tumour-inhibitory effects. To exert its effect, rapamycin binds an immunophilin FK-506-binding protein 12, and this complex inhibits the cellular target of rapamycin, mTOR.
- SERIAL ANALYSIS OF GENE EXPRESSION
-
(SAGE). A technique that is used to identify and quantitate genes that are differentially expressed between two different samples. Sage libraries are generated by adding a short nucleotide sequence tag (10 base pairs) to cDNAs from different RNA samples that are being analysed. The abundance of a given tag within the SAGE library corresponds to the expression level of the corresponding gene, which can easily be identified by sequencing directly from the transcript tag.
Rights and permissions
About this article
Cite this article
Ruggero, D., Pandolfi, P. Does the ribosome translate cancer?. Nat Rev Cancer 3, 179–192 (2003). https://doi.org/10.1038/nrc1015
Issue Date:
DOI: https://doi.org/10.1038/nrc1015