Key Points
-
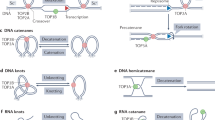
Type II topoisomerases change DNA topology by generating transient DNA double strand breaks and are essential for all eukaryotic cells.
-
Mammalian cells have two topoisomerase II (TOP2) isoforms, TOP2α and TOP2β. TOP2α is essential for all cells, and is essential for separating replicated chromosomes. TOP2β is required for normal development, but is dispensable in some cell types. Type II topoisomerases are required for other processes such as transcription, and the precise roles of the two isoforms in these processes are a subject of current studies.
-
Type II topoisomerases use a two gate mechanism for carrying out topological changes in DNA. The enzyme requires ATP hydrolysis for its reaction. ATP hydrolysis is used for for conformational changes of the enzyme, and is not directly involved in DNA breakage or resealing.
-
Crystal structures of several domains of yeast Top2 have provided additional information about how the enzyme carries out its reactions. A recent structure of the breakage reunion domain of yeast Top2 bound to DNA has shown that the enzyme induces a large bend in the DNA that is cleaved by the enzyme.
-
Biological functions of TOP2 isoforms are modulated by a variety of protein–protein interactions. Some of these interactions may affect enzyme activity, stability and localization.
-
TOP2 activity is also modulated by post-translational modification. In addition to phosphorylation, a crucial post-translational modification of TOP2 is sumoylation. Failure to sumoylate TOP2α or to remove the SUMO modification disrupts the ability of TOP2α to separate replicated chromosomes.
-
TOP2β has a key role in the survival of some neural cells. TOP2β is important in transcriptional regulation, and it is likely that TOP2β enzyme activity is specifically required.
-
Some aspects of TOP2 function during the cell cycle are monitored by checkpoints. It has been hypothesized that a major role of checkpoints is to monitor the completion of decatenation. If so, then TOP2-dependent checkpoints may be crucial for normal chromosome segregation and genome stability.
Abstract
DNA topoisomerases are enzymes that disentangle the topological problems that arise in double-stranded DNA. Many of these can be solved by the generation of either single or double strand breaks. However, where there is a clear requirement to alter DNA topology by introducing transient double strand breaks, only DNA topoisomerase II (TOP2) can carry out this reaction. Extensive biochemical and structural studies have provided detailed models of how TOP2 alters DNA structure, and recent molecular studies have greatly expanded knowledge of the biological contexts in which TOP2 functions, such as DNA replication, transcription and chromosome segregation — processes that are essential for preventing tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001).
Wang, J. C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31, 107–144 (1998).
Sundin, O. & Varshavsky, A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell 25, 659–669 (1981). The first demonstration that replication specifically requires a type II topoisomerase.
Sundin, O. & Varshavsky, A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell 21, 103–114 (1980).
Bates, A. D. & Maxwell, A. DNA topology (Oxford University Press, Oxford, 2005).
Aravind, L., Leipe, D. D. & Koonin, E. V. Toprim — a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26, 4205–4213 (1998).
Classen, S., Olland, S. & Berger, J. M. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl Acad. Sci. USA 100, 10629–10634 (2003). The structure of the ATPase domain of yeast Top2. These authors also reported the structure of the ATPase domain of Top2 bound to a bisdioxopiperazines.
Wei, H., Ruthenburg, A. J., Bechis, S. K. & Verdine, G. L. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 280, 37041–37047 (2005).
Dong, K. C. & Berger, J. M. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450, 1201–1205 (2007). A landmark paper showing the structure of the breakage reunion domain of TOP2 bound to DNA.
Corbett, K. D. & Berger, J. M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004).
Schoeffler, A. J. & Berger, J. M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 (2008). A recent review that integrates structural and biochemical information on different classes of topoisomerases.
Berger, J. M., Gamblin, S. J., Harrison, S. C. & Wang, J. C. Structure and mechanism of DNA topoisomerase II Nature 379, 225–232 (1996); erratum Nature 380,179 (1996). The first structure of the breakage reunion domain of a type II topoisomerase.
Fass, D., Bogden, C. E. & Berger, J. M. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nature Struct. Biol. 6, 322–326 (1999).
Rybenkov, V. V., Ullsperger, C., Vologodskii, A. V. & Cozzarelli, N. R. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science 277, 690–693 (1997). The discovery of the phenomenon of topologoy simplification by type IIA topoisomerases.
Vologodskii, A. V. et al. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl Acad. Sci. USA 98, 3045–3049 (2001).
Chang, C. J., Goulding, S., Earnshaw, W. C. & Carmena, M. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J. Cell Sci. 116, 4715–4726 (2003).
Akimitsu, N. et al. Induction of apoptosis by depletion of DNA topoisomerase II alpha in mammalian cells. Biochem. Biophys. Res. Comm. 307, 301–307 (2003).
Toyoda, Y. & Yanagida, M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell 17, 2287–2302 (2006).
Coelho, P. A. et al. Dual role of Topoisomerase II in centromere resolution and Aurora B activity. Plos Biol. 6, 1758–1777 (2008).
McPherson, J. P. & Goldenberg, G. J. Induction of apoptosis by deregulated expression of DNA topoisomerase IIα. Cancer Res. 58, 4519–4524 (1998).
Mo, Y. Y., Ameiss, K. A. & Beck, W. T. Overexpression of human DNA topoisomerase II alpha by fusion to enhanced green fluorescent protein. Biotechniques 25, 1052–1057 (1998).
Lucas, I., Germe, T., Chevrier-Miller, M. & Hyrien, O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 20, 6509–6519 (2001).
Peter, B. J., Ullsperger, C., Hiasa, H., Marians, K. J. & Cozzarelli, N. R. The structure of supercoiled intermediates in DNA replication. Cell 94, 819–827 (1998). Demonstration that precatenanes were a relevant topological form of replicating DNA.
Postow, L., Crisona, N. J., Peter, B. J., Hardy, C. D. & Cozzarelli, N. R. Topological challenges to DNA replication: Conformations at the fork. Proc. Natl Acad. Sci. USA 98, 8219–8226 (2001).
Brill, S. J., DiNardo, S., Voelkel-Meiman, K. & Sternglanz, R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326, 414–416 (1987); erratum 326, 812 (1987).
Kim, R. A. & Wang, J. C. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 208, 257–267 (1989).
Bermejo, R. et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 21, 1921–1936 (2007).
Holm, C., Goto, T., Wang, J. C. & Botstein, D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell 41, 553–563 (1985). The first analysis of a conditional TOP2 mutant in a eukaryotic cell. This paper confirmed the prediction of Sundin and Varshavsky that TOP2 is required to separate replicated chromosomes.
Baxter, J. & Diffley, J. F. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell 30, 790–802 (2008). An elegant study assessing roles of Top2 in replication in yeast.
Carpenter, A. J. & Porter, A. C. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIα mutant human cell line. Mol. Biol. Cell 15, 5700–5711 (2004). A novel approach to generating a conditional TOP2α allele in a human cell line.
Li, H., Wang, Y. & Liu, X. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J. Biol. Chem. 283, 6209–6221 (2008).
McClendon, A. K., Rodriguez, A. C. & Osheroff, N. Human topoisomerase II α rapidly relaxes positively supercoiled DNA — Implications for enzyme action ahead of replication forks. J. Biol. Chem. 280, 39337–39345 (2005). A demonstration that TOP2α has preferential activity against positively supercoiled DNA. This result may be important for understanding a specific requirement for TOP2α during replication.
Murray, A. W. & Szostak, J. W. Chromosome segregation in mitosis and meiosis. Annu. Rev. Cell Biol. 1, 289–315 (1985).
Losada, A. & Hirano, T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19, 1269–1287 (2005).
Diaz-Martinez, L. A., Gimenez-Abian, J. F. & Clarke, D. J. Chromosome cohesion — rings, knots, orcs and fellowship. J. Cell Sci. 121, 2107–2114 (2008).
Michaelis, C., Ciosk, R. & Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35–45 (1997).
Huang, C. E., Milutinovich, M. & Koshland, D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Phil. Trans. R. Soc. B Biol. Sci. 360, 537–542 (2005).
Bhat, M. A., Philp, A. V., Glover, D. M. & Bellen, H. J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 87, 1103–1114 (1996).
Coelho, P. A., Queiroz-Machado, J. & Sunkel, C. E. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 116, 4763–4776 (2003).
Rattner, J. B., Hendzel, M. J., Furbee, C. S., Muller, M. T. & BazettJones, D. P. Topoisomerase II α is associated with the mammalian centromere in a cell cycle and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 134, 1097–1107 (1996).
Barthelmes, H. U., Grue, P., Feineis, S., Straub, T. & Boege, F. Active DNA topoisomerase II alpha is a component of the salt-stable centrosome core. J. Biol. Chem. 275, 38823–38830 (2000).
Christensen, M. O. et al. Dynamics of human DNA topoisomerases II α and II β in living cells. J. Cell Biol. 157, 31–44 (2002).
Shimogawa, M. M., Widlund, P. O., Riffle, M., Ess, M. & Davis, T. N. Bir1 is required for the tension checkpoint. Mol. Biol. Cell 20, 915–923 (2009).
Liu, D., Vader, G., Vromans, M. J. M., Lampson, M. A. & Lens, S. M. A. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science 323, 1350–1353 (2009).
Huang, H. M. et al. Phosphorylation sites in BubR1 that regulate kinetochore attachment, tension, and mitotic exit. J. Cell Biol. 183, 667–680 (2008).
Porter, A. C. & Farr, C. J. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 12, 569–583 (2004).
Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N. & Elledge, S. J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9, 1169–1182 (2002). The biological significance of the modification of TOP2 with SUMO is described.
Azuma, Y., Arnaoutov, A., Anan, T. & Dasso, M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172–2182 (2005).
Diaz-Martinez, L. A. et al. PIASgamma is required for faithful chromosome segregation in human cells. PLoS ONE 1, e53 (2006).
Roth, W. et al. PIASy-deficient mice display modest defects in IFN and Wnt signaling. J. Immunol. 173, 6189–6199 (2004).
Wong, K. A. et al. Protein inhibitor of activated STAT y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol. Cell. Biol. 24, 5577–5586 (2004).
Dawlaty, M. M. et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIα. Cell 133, 103–115 (2008). Convincing evidence that RANBP2 is required to sumoylate mammalian TOP2.
Belmont, A. S. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 18, 632–638 (2006).
Xu, Y. X. & Manley, J. L. The prolyl isomerase Pin1 functions in mitotic chromosome condensation. Mol. Cell 26, 287–300 (2007).
Maeshima, K. & Laemmli, U. K. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell 4, 467–480 (2003).
Adachi, Y., Kas, E. & Laemmli, U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 8, 3997–4006 (1989).
Gasser, S. M., Laroche, T., Falquet, J., Boy de la Tour, E. & Laemmli, U. K. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 188, 613–629 (1986).
Schultz, M. C., Brill, S. J., Ju, Q., Sternglanz, R. & Reeder, R. H. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 6, 1332–1341 (1992).
Goto, T. & Wang, J. C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I, and construction of mutants defective in both DNA topoisomerase I and DNA topoisomerase II. Proc. Natl Acad. Sci. USA 82, 7178–7182 (1985).
Gartenberg, M. R. & Wang, J. C. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc. Natl Acad. Sci. USA 89, 11461–11465 (1992).
Salceda, J., Fernandez, X. & Roca, J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 25, 2575–2583 (2006).
Mondal, N. & Parvin, J. D. DNA topoisomerase IIα is required for RNA polymerase II transcription on chromatin templates. Nature 413, 435–438 (2001).
Mondal, N. et al. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 31, 5016–5024 (2003).
Kretzschmar, M., Meisterernst, M. & Roeder, R. G. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA 90, 11508–11512 (1993).
Merino, A., Madden, K. R., Lane, W. S., Champoux, J. J. & Reinberg, D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature 365, 227–232 (1993).
Shykind, B. M., Kim, J., Stewart, L., Champoux, J. J. & Sharp, P. A. Topoisomerase I enhances TFIID–TFIIA complex assembly during activation of transcription. Genes Dev. 11, 397–407 (1997).
Liu, L. F. & Wang, J. C. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA 84, 7024–7027 (1987). The transcriptional supercoiling model.
Wang, J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell Biol. 3, 430–440 (2002).
Ju, B. G. & Rosenfeld, M. G. A breaking strategy for topoisomerase IIβ/PARP-1-dependent regulated transcription. Cell Cycle 5, 2557–2560 (2006).
Ju, B. G. et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006). A demonstration that TOP2β has a crucial enzymatic role in transcription. Reference 56 is an important elaboration on the findings of this paper.
McNamara, S., Wang, H., Hanna, N. & Miller, W. H. Jr. Topoisomerase IIβ negatively modulates retinoic acid receptor α function: a novel mechanism of retinoic acid resistance. Mol. Cell. Biol. 28, 2066–2077 (2008).
Lyu, Y. L. et al. Role of topoisomerase IIβ in the expression of developmentally regulated genes. Mol. Cell. Biol. 26, 7929–7941 (2006).
Lyu, Y. L. & Wang, J. C. Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIβ. Proc. Natl Acad. Sci. USA 100, 7123–7128 (2003).
Yang, X., Li, W., Prescott, E. D., Burden, S. J. & Wang, J. C. DNA topoisomerase IIβ and neural development. Science 287, 131–134 (2000).
Hartwell, L. H. & Weinert, T. A. Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634 (1989).
Holm, C., Stearns, T. & Botstein, D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 9, 159–168 (1989).
Uemura, T. & Yanagida, M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 3, 1737–1744 (1984).
Uemura, T. et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50, 917–925 (1987).
Downes, C. S., Mullinger, A. M. & Johnson, R. T. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc. Natl Acad. Sci. USA 88, 8895–8899 (1991).
Clarke, D. J., Johnson, R. T. & Downes, C. S. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J. Cell Sci. 105 (Pt 2), 563–569 (1993).
Clifford, B., Beljin, M., Stark, G. R. & Taylor, W. R. G2 arrest in response to topoisomerase II inhibitors: the role of p53. Cancer Res. 63, 4074–4081 (2003).
Ishida, R. et al. Inhibition of intracellular topoisomerase II by antitumor bis(2,6-dioxopiperazine) derivatives: mode of cell growth inhibition distinct from that of cleavable complex-forming type inhibitors. Cancer Res. 51, 4909–4916 (1991).
Downes, C. S. et al. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature 372, 467–470 (1994). The first paper demonstrating a mitotic delay induced by bisdioxopiperazines. This paper is important in understanding TOP2 checkpoints as it was the first to use a catalytic inhibitor of TOP2 rather than an agent that induces TOP2-mediated DNA damage.
Deming, P. B. et al. The human decatenation checkpoint. Proc. Natl Acad. Sci. USA 98, 12044–12049 (2001).
Skoufias, D. A., Lacroix, F. B., Andreassen, P. R., Wilson, L. & Margolis, R. L. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell 15, 977–990 (2004).
Andrews, C. A. et al. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 20, 1162–1174 (2006).
Damelin, M. & Bestor, T. H. Decatenation checkpoint deficiency destabilizes the stem cell genome. Cell Cycle 5, 345–346 (2006).
Damelin, M., Sun, Y. E., Sodja, V. B. & Bestor, T. H. Decatenation checkpoint deficiency in stem and progenitor cells. Cancer Cell 8, 479–484 (2005).
Luo, K. T., Yuan, J., Chen, J. J. & Lou, Z. K. Topoisomerase II α controls the decatenation checkpoint. Nature Cell Biol. 11, 204–U196 (2009).
Wells, N. J., Addison, C. M., Fry, A. M., Ganapathi, R. & Hickson, I. D. Serine 1524 is a major site of phosphorylation on human topoisomerase II alpha protein in vivo and is a substrate for casein kinase II in vitro. J. Biol. Chem. 269, 29746–29751 (1994).
Wood, J. L. & Chen, J. J. DNA-damage checkpoints: location, location, location. Trends Cell Biol. 18, 451–455 (2008).
Haggarty, S. J. et al. Small molecule modulation of the human chromatid decatenation checkpoint. Chem. Biol. 10, 1267–1279 (2003).
Nitiss, J. L. in DNA Damage and Repair: Volume II: DNA repair in Higher Eukaryotes (eds Nickoloff, J. A. & Hoekstra, M. F.) 517–537 (Humana Press, Totawa, New Jersey, 1998).
Kingma, P. S. & Osheroff, N. The response of eukaryotic topoisomerases to DNA damage. Biochim. Biophys. Acta 1400, 223–232 (1998).
Yamane, K., Wu, X. & Chen, J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22, 555–566 (2002).
Wilstermann, A. M. & Osheroff, N. Base excision repair intermediates as topoisomerase II poisons. J. Biol. Chem. 276, 46290–46296 (2001).
Nitiss, J. L. Targeting DNA topoisomerase II in cancer chemotherapy. Nature Rev. Cancer 8, 338–350 (2008).
Corbett, K. D. & Berger, J. M. Emerging roles for plant topoisomerase VI. Chem. Biol. 10, 107–111 (2003).
Lichten, M. Meiotic recombination: breaking the genome to save it. Curr. Biol. 11, R253–256 (2001).
Austin, C. A. & Marsh, K. L. Eukaryotic DNA topoisomerase II β. Bioessays 20, 215–226 (1998).
Linka, R. M. et al. C-terminal regions of topoisomerase II α and II β determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Res. 35, 3810–3822 (2007).
Keeney, S. et al. A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics 61, 170–182 (1999).
Romanienko, P. J. & Camerini-Otero, R. D. Cloning, characterization, and localization of mouse and human SPO11. Genomics 61, 156–169 (1999).
Sugimoto-Shirasu, K., Stacey, N. J., Corsar, J., Roberts, K. & McCann, M. C. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12, 1782–1786 (2002).
Harkins, T. T. & Lindsley, J. E. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis. Biochemistry 37, 7292–7298 (1998).
Berger, J. M., Gamblin, S. J., Harrison, S. C. & Wang, J. C. Structure and mechanism of DNA topoisomerase II. Nature 379, 225–232 (1996).
Kurz, E. U., Leader, K. B., Kroll, D. J., Clark, M. & Gieseler, F. Modulation of human DNA topoisomerase IIα function by interaction with 14-3-3ɛ. J. Biol. Chem. 275, 13948–13954 (2000).
Wang, Y., Azuma, Y., Moore, D., Osheroff, N. & Neufeld, K. L. Interaction between tumor suppressor adenomatous polyposis coli and topoisomerase II α: implication for the G2/M transition. Mol. Biol. Cell 19, 4076–4085 (2008).
Morrison, C. et al. Proteomic analysis of human metaphase chromosomes reveals topoisomerase II α as an Aurora B substrate. Nucleic Acids Res. 30, 5318–5327 (2002).
Lou, Z., Minter-Dykhouse, K. & Chen, J. BRCA1 participates in DNA decatenation. Nature Struct. Mol. Biol. 12, 589–593 (2005).
Oliveira, R. A., Coelho, P. A. & Sunkel, C. E. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol. 25, 8971–8984 (2005).
Ackerman, P., Glover, C. V. & Osheroff, N. Phosphorylation of DNA topoisomerase II by casein kinase II: modulation of eukaryotic topoisomerase II activity in vitro. Proc. Natl Acad. Sci. USA 82, 3164–3168 (1985).
DeVore, R. F., Corbett, A. H. & Osheroff, N. Phosphorylation of topoisomerase II by casein kinase II and protein kinase C: effects on enzyme-mediated DNA cleavage/religation and sensitivity to the antineoplastic drugs etoposide and 4′-(9-acridinylamino)methane-sulfon-m-anisidide. Cancer Res. 52, 2156–2161 (1992).
Redwood, C., Davies, S. L., Wells, N. J., Fry, A. M. & Hickson, I. D. Casein kinase II stabilizes the activity of human topoisomerase IIα in a phosphorylation-independent manner. J. Biol. Chem. 273, 3635–3642 (1998).
Cardenas, M. E. & Gasser, S. M. Regulation of topoisomerase II by phosphorylation: a role for casein kinase II. J. Cell Sci. 104 (Pt 2), 219–225 (1993).
Isaacs, R. J. et al. Physiological regulation of eukaryotic topoisomerase II. Biochim. Biophys. Acta 1400, 121–137 (1998).
Ahn, B. H., Kim, T. H. & Bae, Y. S. Mapping of the interaction domain of the protein kinase CKII β subunit with target proteins. Mol. Cells 12, 158–163 (2001).
Wells, N. J. & Hickson, I. D. Human topoisomerase II alpha is phosphorylated in a cell-cycle phase-dependent manner by a proline-directed kinase. Eur. J. Biochem. 231, 491–497 (1995).
Poot, R. A. et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19, 3377–3387 (2000).
Varga-Weisz, P. D. et al. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388, 598–602 (1997).
Turner, J. G., Engel, R., Derderian, J. A., Jove, R. & Sullivan, D. M. Human topoisomerase IIα nuclear export is mediated by two CRM-1-dependent nuclear export signals. J. Cell Sci. 117, 3061–3071 (2004).
Mirski, S. E. et al. Topoisomerase II binds importin α isoforms and exportin/CRM1 but does not shuttle between the nucleus and cytoplasm in proliferating cells. Exp. Cell Res. 313, 627–637 (2007).
Tsai, S. C. et al. Histone deacetylase interacts directly with DNA topoisomerase II. Nature Genet. 26, 349–353 (2000).
Johnson, C. A., Padget, K., Austin, C. A. & Turner, B. M. Deacetylase activity associates with topoisomerase II and is necessary for etoposide-induced apoptosis. J. Biol. Chem. 276, 4539–4542 (2001).
Yun, J., Tomida, A., Andoh, T. & Tsuruo, T. Interaction between glucose-regulated destruction domain of DNA topoisomerase IIα and MPN domain of Jab1/CSN5. J. Biol. Chem. 279, 31296–31303 (2004).
Matheos, D., Ruiz, M. T., Price, G. B. & Zannis-Hadjopoulos, M. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochim. Biophys. Acta 1578, 59–72 (2002).
Cowell, I. G. et al. Human topoisomerase IIα and IIβ interact with the C-terminal region of p53. Exp. Cell Res. 255, 86–94 (2000).
Niimi, A., Suka, N., Harata, M., Kikuchi, A. & Mizuno, S. Co-localization of chicken DNA topoisomerase IIα, but not β, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma 110, 102–114 (2001).
Messenger, M. M. et al. Interactions between protein kinase CK2 and Pin1. Evidence for phosphorylation-dependent interactions. J. Biol. Chem. 277, 23054–23064 (2002).
Cuvier, O., Stanojcic, S., Lemaitre, J. M. & Mechali, M. A topoisomerase II-dependent mechanism for resetting replicons at the S–M-phase transition. Genes Dev. 22, 860–865 (2008).
Wyles, J. P., Wu, Z., Mirski, S. E. & Cole, S. P. Nuclear interactions of topoisomerase II α and β with phospholipid scramblase 1. Nucleic Acids Res. 35, 4076–4085 (2007).
Navarro, M. S. & Bachant, J. RanBP2: a tumor suppressor with a new twist on TopoII, SUMO, and centromeres. Cancer Cell 13, 293–295 (2008).
Zhou, K. et al. RNA helicase A interacts with dsDNA and topoisomerase IIalpha. Nucleic Acids Res. 31, 2253–2260 (2003).
Mao, Y., Desai, S. D. & Liu, L. F. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. 275, 26066–26073 (2000).
Agostinho, M. et al. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 68, 2409–2418 (2008).
Takahashi, Y. & Strunnikov, A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma 117, 189–198 (2008).
Huang, L. et al. Functional interaction of DNA topoisomerase II α with the β-catenin and T-cell factor-4 complex. Gastroenterology 133, 1569–1578 (2007).
Yamane, K., Kawabata, M. & Tsuruo, T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 250, 794–799 (1997).
Lee, C. G., Hague, L. K., Li, H. & Donnelly, R. Identification of toposome, a novel multisubunit complex containing topoisomerase IIα. Cell Cycle 3, 638–647 (2004).
Acknowledgements
The author thanks J. Berger, University of California Berkeley, USA, who kindly provided figures and also provided useful discussion, and also Y. Pommier, National Cancer Institute, Bethesda, for encouragement. Work in the author's laboratory is supported by grants from the National Cancer Institute (CA82313 and CA21765) and the American Lebanese Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Glossary
- Catenane
-
Circles linked as in a chain: the two links cannot be separated without breaking one of the two molecules.
- TOPRIM domain
-
A conserved domain found in topoisomerases, primases and other DNA metabolic enzymes. The TOPRIM domain adopts a Rossman fold and is involved in divalent cation binding.
- B-DNA
-
DNA exists in many possible conformations, but only A-DNA, B-DNA and Z-DNA have been observed in organisms. Which conformation DNA adopts depends, for example, on the sequence of the DNA, or the amount and direction of supercoiling. The B form is most common under the conditions found in cells.
- Boltzmann distribution
-
A certain distribution function or probability measure for the distribution of the states of a system.
- Precatenane
-
A structure related to a catenane that results from the interwinding of DNA strands behind a replication fork. Precatenanes interconvert with positive supercoils that arise in front of a replication fork.
- Hypomorphs
-
Organisms expressing alleles that result in a reduction, but not the elimination, of wild-type levels of a gene product or activity, often causing a less severe phenotype than a loss-of-function (or null) allele.
- Bisdioxopiperazines
-
A class of small molecules, including ICRF-159, ICRF-187 and MST-16, that inhibit the catalytic activity of TOP2 and do not stabilize the TOP2 cleaved complex. Bisdioxopiperazines are the most commonly used catalytic inhibitors of type II topoisomerases.
Rights and permissions
About this article
Cite this article
Nitiss, J. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer 9, 327–337 (2009). https://doi.org/10.1038/nrc2608
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2608
This article is cited by
-
The role of H3K27me3 methylation in cancer development
Genome Instability & Disease (2024)
-
A sumoylation program is essential for maintaining the mitotic fidelity in proliferating mantle cell lymphoma cells
Experimental Hematology & Oncology (2022)
-
Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions
Nature Communications (2022)
-
Three-dimensional quantitative structural-activity relationship and molecular dynamics study of multivariate substituted 4-oxyquinazoline HDAC6 inhibitors
Molecular Diversity (2022)
-
Aconitine linoleate, a natural lipo-diterpenoid alkaloid, stimulates anti-proliferative activity reversing doxorubicin resistance in MCF-7/ADR breast cancer cells as a selective topoisomerase IIα inhibitor
Naunyn-Schmiedeberg's Archives of Pharmacology (2022)