Key Points
-
Pim kinases are frequently overexpressed in human haematological malignancies and solid cancers, and they are often associated with strongly elevated MYC levels.
-
Overexpression of Pim kinases is associated with a good prognosis in some solid cancers, such as prostate cancer, but is associated with a poor prognosis in other solid cancers and most haematological malignancies.
-
Pim kinases are serine/threonine kinases that have consititutive activity (and therefore lack the need for post-translational activation). As their mRNA and proteins have a very short half-life, the activity of Pim kinases is largely regulated at the transcriptional and translational levels.
-
Pim kinases mediate survival signalling through phosphorylation of BCL-2-associated agonist of cell death (BAD), which induces release of the anti-apoptotic BCL-2 and BCL-2-like 1 (also known as BCL-X) proteins and thus lowers the threshold for apoptosis. Pim kinases might also induce BAD activities towards the regulation of glucose metabolism.
-
PIM2 can regulate cap-dependent translation in a mammalian target of rapamycin complex 1 (mTORC1)-independent manner, and in parallel to the PI3K–Akt pathway. This activity has been found to be relevant for certain human haematological malignancies, and is not shared with PIM1 or PIM3.
-
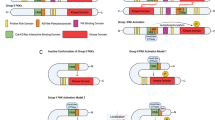
PIM1 can bind to MYC–MAX complexes and phosphorylate H3S10 at E-boxes, thereby setting off a cascade of events that leads to transcriptional pause release of RNA polymerase II at MYC-driven promoters. It remains unknown whether this activity contributes to tumorigenesis in vivo or whether other Pim family members share this activity with PIM1.
-
Pim kinases are promising targets for pharmacological inhibition, as the structural conformation of the ATP-binding pocket in the active site is different from that of other protein kinases, which in theory should allow the design of specific and selective inhibitors. The lack of any overt phenotypes in Pim1−/−;Pim2−/−;Pim3−/− mice indicates that such drugs might have a low toxicity profile.
Abstract
Pim oncogenes are overexpressed in a wide range of tumours from a haematological and epithelial origin. Pim genes encode serine/threonine kinases that have been shown to counteract the increased sensitivity to apoptosis induction that is associated with MYC-driven tumorigenesis. Recently, considerable progress has been made in characterizing the pathways of PIM-mediated survival signalling. Given the unique structure of their active site and the minimal phenotype of mice mutant for all Pim family members, these oncogenes might be promising targets for highly specific and selective drugs with favourable toxicity profiles. In this Review, we discuss the physiological functions and oncogenic activities of Pim kinases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cuypers, H. T. et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell 37, 141–150 (1984).
Selten, G. et al. The primary structure of the putative oncogene pim-1 shows extensive homology with protein kinases. Cell 46, 603–611 (1986).
Hoover, D., Friedmann, M., Reeves, R. & Magnuson, N. S. Recombinant human pim-1 protein exhibits serine/threonine kinase activity. J. Biol. Chem. 266, 14018–14023 (1991).
Padma, R. & Nagarajan, L. The human PIM-1 gene product is a protein serine kinase. Cancer Res. 51, 2486–2489 (1991).
Saris, C. J., Domen, J. & Berns, A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 10, 655–664 (1991).
van Lohuizen, M. et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56, 673–682 (1989).
Selten, G., Cuypers, H. T., Zijlstra, M., Melief, C. & Berns, A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 3, 3215–3222 (1984).
Verbeek, S. et al. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol. Cell. Biol. 11, 1176–1179 (1991).
Mikkers, H. et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nature Genet. 32, 153–159 (2002).
van der Lugt, N. M. et al. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J. 14, 2536–2544 (1995).
Fox, C. J. et al. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 17, 1841–1854 (2003). This paper is the first study to describe two novel roles for Pim kinases in mediating cell survival: PIM2-specific regulation of 4E-BP1 and the phosphorylation of BAD by PIM2, which was later observed to be shared with the other family members.
Qian, K. C. et al. Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J. Biol. Chem. 280, 6130–6137 (2005).
Laird, P. W. et al. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 21, 4750–4755 (1993).
Domen, J. et al. Impaired interleukin-3 response in Pim-1-deficient bone marrow-derived mast cells. Blood 82, 1445–1452 (1993).
Mikkers, H. et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol. Cell. Biol. 24, 6104–6115 (2004). This study describes the phenotype of Pim2−/− and Pim3−/− mice, as well as that of the Pim1−/−;Pim2−/−;Pim3−/− mouse, and finds that Pim kinases are not required for crucial developmental processes.
Feldman, J. D. et al. KID-1, a protein kinase induced by depolarization in brain. J. Biol. Chem. 273, 16535–16543 (1998).
Konietzko, U. et al. Pim kinase expression is induced by LTP stimulation and required for the consolidation of enduring LTP. EMBO J. 18, 3359–3369 (1999).
Katakami, N. et al. Role of pim-1 in smooth muscle cell proliferation. J. Biol. Chem. 279, 54742–54749 (2004).
Muraski, J. A. et al. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nature Med. 13, 1467–1475 (2007).
Zippo, A., De, R. A., Bardelli, M., Galvagni, F. & Oliviero, S. Identification of Flk-1 target genes in vasculogenesis: Pim-1 is required for endothelial and mural cell differentiation in vitro. Blood 103, 4536–4544 (2004).
Stewart, B. E. & Rice, R. H. Differentiation-associated expression of the proto-oncogene pim-1 in cultured human keratinocytes. J. Invest. Dermatol. 105, 699–703 (1995).
Gapter, L. A., Magnuson, N. S., Ng, K. Y. & Hosick, H. L. Pim-1 kinase expression during murine mammary development. Biochem. Biophys. Res. Commun. 345, 989–997 (2006).
Domen, J. et al. Pim-1 levels determine the size of early B lymphoid compartments in bone marrow. J. Exp. Med. 178, 1665–1673 (1993).
Cottage, C. T. et al. Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ. Res. 106, 891–901 (2010).
Aksoy, I. et al. Self-renewal of murine embryonic stem cells is supported by the serine/threonine kinases Pim-1 and Pim-3. Stem Cells 25, 2996–3004 (2007).
Miura, O. et al. Induction of tyrosine phosphorylation of Vav and expression of Pim-1 correlates with Jak2-mediated growth signaling from the erythropoietin receptor. Blood 84, 4135–4141 (1994).
Matikainen, S. et al. Interferon-α activates multiple STAT proteins and upregulates proliferation-associated IL-2Rα, c-myc, and pim-1 genes in human T cells. Blood 93, 1980–1991 (1999).
Shirogane, T. et al. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 11, 709–719 (1999).
Castro, A., Sengupta, T. K., Ruiz, D. C., Yang, E. & Ivashkiv, L. B. IL-4 selectively inhibits IL-2-triggered Stat5 activation, but not proliferation, in human T cells. J. Immunol. 162, 1261–1269 (1999).
Wierenga, A. T., Vellenga, E. & Schuringa, J. J. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol. Cell. Biol. 28, 6668–6680 (2008).
Li, J. et al. Novel NEMO/IκB kinase and NF-κB target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276, 18579–18590 (2001).
Zhu, N. et al. CD40 signaling in B cells regulates the expression of the Pim-1 kinase via the NF-κB pathway. J. Immunol. 168, 744–754 (2002).
Elvidge, G. P. et al. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1α, HIF-2α, and other pathways. J. Biol. Chem. 281, 15215–15226 (2006).
Chen, J. et al. Pim-1 plays a pivotal role in hypoxia-induced chemoresistance. Oncogene 28, 2581–2592 (2009).
Zhao, Y. et al. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene 27, 1–8 (2008).
Domen, J. et al. Comparison of the human and mouse PIM-1 cDNAs: nucleotide sequence and immunological identification of the in vitro synthesized PIM-1 protein. Oncogene Res. 1, 103–112 (1987).
Selten, G., Cuypers, H. T. & Berns, A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 4, 1793–1798 (1985).
De Benedetti, A. & Graff, J. R. eIF-4E expression and its role in malignancies and metastases. Oncogene 23, 3189–3199 (2004).
Hoover, D. S., Wingett, D. G., Zhang, J., Reeves, R. & Magnuson, N. S. Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor elF-4E. Cell Growth Differ. 8, 1371–1380 (1997).
Culjkovic, B., Topisirovic, I., Skrabanek, L., Ruiz-Gutierrez, M. & Borden, K. L. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol. 175, 415–426 (2006). This paper characterizes a novel function for EIF4E in regulating nuclear export of mRNA transcripts, based on the recognition of a structural motif in the UTR sequences, and resulting in higher levels of protein translation.
Lilly, M., Sandholm, J., Cooper, J. J., Koskinen, P. J. & Kraft, A. The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 18, 4022–4031 (1999).
Pircher, T. J., Zhao, S., Geiger, J. N., Joneja, B. & Wojchowski, D. M. Pim-1 kinase protects hematopoietic FDC cells from genotoxin-induced death. Oncogene 19, 3684–3692 (2000).
Xie, Y. et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene 25, 70–78 (2006).
Nasser, M. W. et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 283, 33394–33405 (2008).
Eiring, A. M. et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140, 652–665 (2010).
Kim, O. et al. Synergism of cytoplasmic kinases in IL6-induced ligand-independent activation of androgen receptor in prostate cancer cells. Oncogene 23, 1838–1844 (2004).
Bullock, A. N., Debreczeni, J., Amos, A. L., Knapp, S. & Turk, B. E. Structure and substrate specificity of the Pim-1 kinase. J. Biol. Chem. 280, 41675–41682 (2005).
Losman, J. A., Chen, X. P., Vuong, B. Q., Fay, S. & Rothman, P. B. Protein phosphatase 2A regulates the stability of Pim protein kinases. J. Biol. Chem. 278, 4800–4805 (2003).
Ma, J., Arnold, H. K., Lilly, M. B., Sears, R. C. & Kraft, A. S. Negative regulation of Pim-1 protein kinase levels by the B56β subunit of PP2A. Oncogene 26, 5145–5153 (2007).
Mizuno, K. et al. Regulation of Pim-1 by Hsp90. Biochem. Biophys. Res. Commun. 281, 663–669 (2001).
Shay, K. P., Wang, Z., Xing, P. X., McKenzie, I. F. & Magnuson, N. S. Pim-1 kinase stability is regulated by heat shock proteins and the ubiquitin-proteasome pathway. Mol. Cancer Res. 3, 170–181 (2005).
Chen, J. et al. Hypoxia-mediated up-regulation of Pim-1 contributes to solid tumor formation. Am. J. Pathol. 175, 400–411 (2009).
Grundler, R. et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J. Exp. Med. 206, 1957–1970 (2009).
Fox, C. J., Hammerman, P. S. & Thompson, C. B. The Pim kinases control rapamycin-resistant T cell survival and activation. J. Exp. Med. 201, 259–266 (2005).
Schmidt, T. et al. Evidence implicating Gfi-1 and Pim-1 in pre-T-cell differentiation steps associated with β-selection. EMBO J. 17, 5349–5359 (1998).
Jacobs, H. et al. PIM1 reconstitutes thymus cellularity in interleukin 7- and common γ chain-mutant mice and permits thymocyte maturation in Rag- but not CD3γ-deficient mice. J. Exp. Med. 190, 1059–1068 (1999).
Leduc, I. et al. The Pim-1 kinase stimulates maturation of TCRβ-deficient T cell progenitors: implications for the mechanism of Pim-1 action. Int. Immunol. 12, 1389–1396 (2000).
Pearson, R. & Weston, K. c-Myb regulates the proliferation of immature thymocytes following β-selection. EMBO J. 19, 6112–6120 (2000).
Dose, M. et al. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood 108, 2669–2677 (2006).
Zhang, Y., Wang, Z., Li, X. & Magnuson, N. S. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene 27, 4809–4819 (2008).
Winn, L. M., Lei, W. & Ness, S. A. Pim-1 phosphorylates the DNA binding domain of c-Myb. Cell Cycle 2, 258–262 (2003).
Aho, T. L., Sandholm, J., Peltola, K. J., Ito, Y. & Koskinen, P. J. Pim-1 kinase phosphorylates RUNX family transcription factors and enhances their activity. BMC Cell Biol. 7, 21 (2006).
Wang, Z. et al. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim. Biophys. Acta 1593, 45–55 (2002).
Morishita, D., Katayama, R., Sekimizu, K., Tsuruo, T. & Fujita, N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 68, 5076–5085 (2008).
Mochizuki, T. et al. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J. Biol. Chem. 274, 18659–18666 (1999).
Bachmann, M. et al. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int. J. Biochem. Cell Biol. 38, 430–443 (2006).
Chen, X. P. et al. Pim serine/threonine kinases regulate the stability of Socs-1 protein. Proc. Natl Acad. Sci. USA 99, 2175–2180 (2002).
Peltola, K. J. et al. Pim-1 kinase inhibits STAT5-dependent transcription via its interactions with SOCS1 and SOCS3. Blood 103, 3744–3750 (2004).
Gu, J. J., Wang, Z., Reeves, R. & Magnuson, N. S. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene 28, 4261–4271 (2009).
Peng, C. et al. Pim kinase substrate identification and specificity. J. Biochem. 141, 353–362 (2007).
Aho, T. L. et al. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 571, 43–49 (2004).
Macdonald, A. et al. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 7, 1 (2006).
Yan, B. et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J. Biol. Chem. 278, 45358–45367 (2003).
Brault, L. et al. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica 95, 1004–1015 (2010).
Amaravadi, R. & Thompson, C. B. The survival kinases Akt and Pim as potential pharmacological targets. J. Clin. Invest. 115, 2618–2624 (2005).
Choudhary, C. et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell 36, 326–339 (2009).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Wright, G. et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl Acad. Sci. USA 100, 9991–9996 (2003).
Poulsen, C. B. et al. Microarray-based classification of diffuse large B-cell lymphoma. Eur. J. Haematol. 74, 453–465 (2005).
Hsi, E. D. et al. Ki67 and PIM1 expression predict outcome in mantle cell lymphoma treated with high dose therapy, stem cell transplantation and rituximab: a Cancer and Leukemia Group B 59909 correlative science study. Leuk. Lymphoma 49, 2081–2090 (2008).
Dhanasekaran, S. M. et al. Delineation of prognostic biomarkers in prostate cancer. Nature 412, 822–826 (2001). This paper is the first description of a dysregulated expression of Pim kinases in epithelial tumours. PIM1 was specifically overexpressed in prostate tumour samples compared with adjacent normal tissue and benign lesions. Remarkably, PIM1 expression correlated with a good prognosis, which was later confirmed by reference 82.
Rhodes, D. R., Sanda, M. G., Otte, A. P., Chinnaiyan, A. M. & Rubin, M. A. Multiplex biomarker approach for determining risk of prostate-specific antigen-defined recurrence of prostate cancer. J. Natl. Cancer Inst. 95, 661–668 (2003).
Reiser-Erkan, C. et al. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 7, 1352–1359 (2008).
Warnecke-Eberz, U. et al. Frequent down-regulation of pim-1 mRNA expression in non-small cell lung cancer is associated with lymph node metastases. Oncol. Rep. 20, 619–624 (2008).
Warnecke-Eberz, U. et al. Prognostic impact of protein overexpression of the proto-oncogene PIM-1 in gastric cancer. Anticancer Res. 29, 4451–4455 (2009).
Peltola, K. et al. Pim-1 kinase expression predicts radiation response in squamocellular carcinoma of head and neck and is under the control of epidermal growth factor receptor. Neoplasia 11, 629–636 (2009).
Amson, R. et al. The human protooncogene product p33pim is expressed during fetal hematopoiesis and in diverse leukemias. Proc. Natl Acad. Sci. USA 86, 8857–8861 (1989).
von Lindern, M., van Agthoven, T., Hagemeijer, A., Adriaansen, H. & Grosveld, G. The human pim-1 gene is not directly activated by the translocation (6;9) in acute nonlymphocytic leukemia. Oncogene 4, 75–79 (1989).
Sivertsen, E. A. et al. Gain of chromosome 6p is an infrequent cause of increased PIM1 expression in B-cell non-Hodgkin's lymphomas. Leukemia 20, 539–542 (2006).
Dave, S. S. et al. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 354, 2431–2442 (2006).
Hammerman, P. S. et al. Lymphocyte transformation by Pim-2 is dependent on nuclear factor-κB activation. Cancer Res. 64, 8341–8348 (2004).
Davis, R. E., Brown, K. D., Siebenlist, U. & Staudt, L. M. Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 194, 1861–1874 (2001).
Pasqualucci, L. et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412, 341–346 (2001).
Deutsch, A. J., Fruhwirth, M., Aigelsreiter, A., Cerroni, L. & Neumeister, P. Primary cutaneous marginal zone B-cell lymphomas are targeted by aberrant somatic hypermutation. J. Invest. Dermatol. 129, 476–479 (2009).
Halldorsdottir, A. M. et al. Quantifying the role of aberrant somatic hypermutation in transformation of follicular lymphoma. Leuk. Res. 32, 1015–1021 (2008).
Deutsch, A. J. et al. MALT lymphoma and extranodal diffuse large B-cell lymphoma are targeted by aberrant somatic hypermutation. Blood 109, 3500–3504 (2007).
Liso, A. et al. Aberrant somatic hypermutation in tumor cells of nodular-lymphocyte-predominant and classic Hodgkin lymphoma. Blood 108, 1013–1020 (2006).
Rossi, D. et al. Aberrant somatic hypermutation in transformation of follicular lymphoma and chronic lymphocytic leukemia to diffuse large B-cell lymphoma. Haematologica 91, 1405–1409 (2006).
Hemann, M. T. et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436, 807–811 (2005).
Dang, C. V., O'donnell, K. A. & Juopperi, T. The great MYC escape in tumorigenesis. Cancer Cell 8, 177–178 (2005).
Eilers, M. & Eisenman, R. N. Myc's broad reach. Genes Dev. 22, 2755–2766 (2008).
Fernandez, P. C. et al. Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003).
Hargreaves, D. C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009). This paper dissects in detail the differential regulation of the induction of gene expression between primary and secondary response genes, and identified trancriptional pause release as an important regulatory mechanism.
Rahl, P. B. et al. c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010). This paper characterizes transcriptional pause release as the major mechanism of the induction of gene expression by Myc in murine ES cells.
Zippo, A., De, R. A., Serafini, R. & Oliviero, S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nature Cell Biol. 9, 932–944 (2007).
Winter, S. et al. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 27, 88–99 (2008).
Zippo, A. et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136 (2009). References 105 and 107 characterize an entirely new function for PIM1 kinase in the phosphorylation of H3S10 in the promoter regions of MYC target genes, facilitating transcriptional elongation at these loci and contributing to the MYC-driven transcriptional programme.
Naud, J. F. & Eilers, M. PIM1 and MYC: a changing relationship? Nature Cell Biol. 9, 873–875 (2007).
Hammerman, P. S., Fox, C. J., Birnbaum, M. J. & Thompson, C. B. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood 105, 4477–4483 (2005).
Woodland, R. T. et al. Multiple signaling pathways promote B lymphocyte stimulator dependent B-cell growth and survival. Blood 111, 750–760 (2008).
Tamburini, J. et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood 114, 1618–1627 (2009). This paper shows that PIM2-dependent regulation of cap-dependent translation was crucial for rapamycin-insensitive deregulation of oncogenic protein synthesis in AML blast cells, and implicates the regulation of protein translation as a relevant therapeutic target.
Bordeleau, M. E. et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 118, 2651–2660 (2008).
Hsieh, A. C. et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 17, 249–261 (2010). This paper extends the findings in reference 111 by showing that mTOR active site-specific inhibitors induce a response in rapamycin-insensitive tumours, in a 4EBP1- and EIF4E-dependent manner and resulting in downregulation of MCL1 expression, implicating mTORC1 as a relevant target for the treatment of human cancers.
Mills, J. R. et al. mTORC1 promotes survival through translational control of Mcl-1. Proc. Natl Acad. Sci. USA 105, 10853–10858 (2008).
Robert, F. et al. Altering chemosensitivity by modulating translation elongation. PLoS ONE 4, e5428 (2009).
Wendel, H. G. et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428, 332–337 (2004).
Chen, L. S., Redkar, S., Bearss, D., Wierda, W. G. & Gandhi, V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood 114, 4150–4157 (2009).
Stewart, M. et al. Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res. 67, 5126–5133 (2007).
Fischer, K. M. et al. Enhancement of myocardial regeneration through genetic engineering of cardiac progenitor cells expressing Pim-1 kinase. Circulation 120, 2077–2087 (2009).
Wu, Y. et al. Accelerated hepatocellular carcinoma development in mice expressing the Pim-3 transgene selectively in the liver. Oncogene 29, 2228–2237 (2010).
Zhang, Y., Wang, Z. & Magnuson, N. S. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol. Cancer Res. 5, 909–922 (2007).
Lin, Y. W. et al. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood 115, 824–833 (2010).
Beharry, Z. et al. Novel benzylidene-thiazolidine-2,4-diones inhibit Pim protein kinase activity and induce cell cycle arrest in leukemia and prostate cancer cells. Mol. Cancer Ther. 8, 1473–1483 (2009).
Zhang, F. et al. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol. Ther. 8, 846–853 (2009).
Bachmann, M., Hennemann, H., Xing, P. X., Hoffmann, I. & Moroy, T. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J. Biol. Chem. 279, 48319–48328 (2004).
Danial, N. N. BAD: undertaker by night, candyman by day. Oncogene 27 (Suppl. 1), S53–S70 (2008).
Li, Y. Y. et al. Pim-3, a proto-oncogene with serine/threonine kinase activity, is aberrantly expressed in human pancreatic cancer and phosphorylates bad to block bad-mediated apoptosis in human pancreatic cancer cell lines. Cancer Res. 66, 6741–6747 (2006).
Zha, J., Harada, H., Yang, E., Jockel, J. & Korsmeyer, S. J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL . Cell 87, 619–628 (1996).
Chiang, C. W. et al. Protein phosphatase 2A dephosphorylation of phosphoserine 112 plays the gatekeeper role for BAD-mediated apoptosis. Mol. Cell. Biol. 23, 6350–6362 (2003).
Danial, N. N. et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424, 952–956 (2003). This paper is the first description of the regulatory role for mitochondrially located BAD on glycolysis and glucose metabolism in hepatocytes from BAD-mutant mice.
Djouder, N. et al. S6K1-mediated disassembly of mitochondrial URI/PP1γ complexes activates a negative feedback program that counters S6K1 survival signaling. Mol. Cell 28, 28–40 (2007).
Danial, N. N. et al. Dual role of proapoptotic BAD in insulin secretion and β cell survival. Nature Med. 14, 144–153 (2008).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Deberardinis, R. J., Lum, J. J., Hatzivassiliou, G. & Thompson, C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 (2008).
Deberardinis, R. J. & Cheng, T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 (2010).
Gao, P. et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009).
Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA 105, 18782–18787 (2008).
Yuneva, M., Zamboni, N., Oefner, P., Sachidanandam, R. & Lazebnik, Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 178, 93–105 (2007).
Li, Y. Y., Wu, Y., Tsuneyama, K., Baba, T. & Mukaida, N. Essential contribution of Ets-1 to constitutive Pim-3 expression in human pancreatic cancer cells. Cancer Sci. 100, 396–404 (2009).
Behan, J. W. et al. Adipocytes impair leukemia treatment in mice. Cancer Res. 69, 7867–7874 (2009).
Mumenthaler, S. M. et al. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Mol. Cancer Ther. 8, 2882–2893 (2009).
Zemskova, M., Sahakian, E., Bashkirova, S. & Lilly, M. The PIM1 kinase is a critical component of a survival pathway activated by docetaxel and promotes survival of docetaxel-treated prostate cancer cells. J. Biol. Chem. 283, 20635–20644 (2008).
Jacobs, M. D. et al. Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002. J. Biol. Chem. 280, 13728–13734 (2005).
Kumar, A. et al. Crystal structures of proto-oncogene kinase Pim1: a target of aberrant somatic hypermutations in diffuse large cell lymphoma. J. Mol. Biol. 348, 183–193 (2005).
Bullock, A. N. et al. Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in moloney murine leukemia virus (PIM-1) kinase. J. Med. Chem. 48, 7604–7614 (2005).
Debreczeni, J. E. et al. Ruthenium half-sandwich complexes bound to protein kinase Pim-1. Angew. Chem. Int. Ed. Engl. 45, 1580–1585 (2006).
Morwick, T. Pim kinase inhibitors: a survey of the patent literature. Expert. Opin. Ther. Pat. 20, 193–212 (2010).
Pogacic, V. et al. Structural analysis identifies imidazo[1,2-b]pyridazines as PIM kinase inhibitors with in vitro antileukemic activity. Cancer Res. 67, 6916–6924 (2007).
Xia, Z. et al. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2 protein kinases. J. Med. Chem. 52, 74–86 (2009).
Ellwood-Yen, K. et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003). This paper describes a mouse model for Myc-driven prostate cancer, and finds PIM1 to be the most consistently regulated gene within a gene signature specific for MYC-driven prostate tumours, in both mice and humans.
Wang, J. et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene 29, 2477–2487 (2010).
Kool, J. & Berns, A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nature Rev. Cancer 9, 389–399 (2009).
Price, D. H. Regulation of RNA polymerase II elongation by c-Myc. Cell 141, 399–400 (2010).
Fuda, N. J., Ardehali, M. B. & Lis, J. T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 (2009).
Price, D. H. Poised polymerases: on your mark...get set...go! Mol. Cell 30, 7–10 (2008).
Chiang, C. W. et al. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin- 3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation. Blood 97, 1289–1297 (2001).
Wang, H. G. et al. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 284, 339–343 (1999).
Jaffe, E. S., Harris, N. L., Stein, H. & Isaacson, P. G. Classification of lymphoid neoplasms: the microscope as a tool for disease discovery. Blood 112, 4384–4399 (2008).
Cohen, A. M. et al. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk. Lymphoma 45, 951–955 (2004).
de Vos, S. et al. Cell cycle alterations in the blastoid variant of mantle cell lymphoma (MCL-BV) as detected by gene expression profiling of mantle cell lymphoma (MCL) and MCL-BV. Diagn. Mol. Pathol. 12, 35–43 (2003).
Zhu, Y. et al. Investigatory and analytical approaches to differential gene expression profiling in mantle cell lymphoma. Br. J. Haematol. 119, 905–915 (2002).
Martelli, M., Ferreri, A. J. & Johnson, P. Primary mediastinal large B-cell lymphoma. Crit. Rev. Oncol. Hematol. 68, 256–263 (2008).
Valdman, A., Fang, X., Pang, S. T., Ekman, P. & Egevad, L. Pim-1 expression in prostatic intraepithelial neoplasia and human prostate cancer. Prostate 60, 367–371 (2004).
Xu, Y. et al. Overexpression of PIM-1 is a potential biomarker in prostate carcinoma. J. Surg. Oncol. 92, 326–330 (2005).
Cibull, T. L. et al. Overexpression of Pim-1 during progression of prostatic adenocarcinoma. J. Clin. Pathol. 59, 285–288 (2006).
Ayala, G. E. et al. Growth and survival mechanisms associated with perineural invasion in prostate cancer. Cancer Res. 64, 6082–6090 (2004).
Dai, H. et al. Pim-2 upregulation: biological implications associated with disease progression and perinueral invasion in prostate cancer. Prostate 65, 276–286 (2005).
Zheng, H. C. et al. Aberrant Pim-3 expression is involved in gastric adenoma-adenocarcinoma sequence and cancer progression. J. Cancer Res. Clin. Oncol. 134, 481–488 (2008).
Chen, C. N. et al. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J. Clin. Oncol. 23, 7286–7295 (2005).
Beier, U. H., Weise, J. B., Laudien, M., Sauerwein, H. & Gorogh, T. Overexpression of Pim-1 in head and neck squamous cell carcinomas. Int. J. Oncol. 30, 1381–1387 (2007).
Chiang, W. F. et al. Up-regulation of a serine-threonine kinase proto-oncogene Pim-1 in oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 35, 740–745 (2006).
Popivanova, B. K. et al. Proto-oncogene, Pim-3 with serine/threonine kinase activity, is aberrantly expressed in human colon cancer cells and can prevent Bad-mediated apoptosis. Cancer Sci. 98, 321–328 (2007).
Gong, J. et al. Serine/threonine kinase Pim-2 promotes liver tumorigenesis induction through mediating survival and preventing apoptosis of liver cell. J. Surg. Res. 153, 17–22 (2009).
Peperzak, V., Veraar, E. A., Keller, A. M., Xiao, Y. & Borst, J. The Pim kinase pathway contributes to survival signaling in primed CD8+ T cells upon CD27 costimulation. J. Immunol. 185, 6670–6678 (2010).
Vlacich, G., Nawijn, M. C., Webb, G. C. & Steiner, D. F. Pim3 negatively regulates glucose-stimulated insulin secretion. Islets 2, 308–317 (2010).
Acknowledgements
The authors would like to thank R. van Amerongen for critical reading of the manuscript. The authors regret not being able to cite all relevant references owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Long terminal repeat
-
LTR. Identical sequences at both ends of the proviral genome comprised of enhancer elements (U3 region), a promoter (U3/R boundary) and a polyadenylation signal (R/U5 boundary). Facilitates proviral integration and regulates expression of viral mRNA and so the replication the virus.
- Pre-B cell lymphomas
-
Hyperplastic growth and transformation of large pre-B cells owing to the accumulation of mutations that affect their ability to keep pre-B cell receptor and MYC levels at bay.
- Kozak sequence
-
The consensus gccRccAUGG (R represents a purine) sequence in the 5′ region of eukaryotic mRNA that is the optimal ribosomal-binding site and facilitates initiation of translation.
- Primary response gene
-
PRG. Upon mitogenic stimuli, upregulation of PRG transcription is rapid but transient and does not require de novo protein synthesis. Most PRGs are transcription factors and propagate signalling by regulating expression of a cascade of downstream secondary target genes.
- Cap-dependent translation
-
Recognition of precursor mRNA transcripts containing a 7-methylguanosine cap (m7G) at their 5′ end and assembly of 40S ribosomal subunit with the help of eukaryotic initiation factors, after which 40S subunit scans towards the 3′ end of mRNA till the first start codon (AUG).
- 5′-m7G cap
-
A post-transcriptional modification of the first 5′ nucleotide of eukaryotic mRNA forming a m7G(5′)ppp(5′)N cap. Protects mRNA from degradation, binds EIF4E, and recruits eIF4F and 40S ribosome subunit to initiate translation.
- Stem-loop-pair
-
Structurally conserved element formed by a set of A and U nucleotides (UX2UX2A) in the eIF4E-sensitive element (4E-SE) of the 3′ UTR. Binds to EIF4E and promotes nuclear export of the mRNA.
- β-selection
-
A positive selection event during the maturation of precursor T cells upon successful rearrangement of the T cell receptor (TCR) β-chain locus that allows pairing to the pre-Tα chain to form a functional pre-TCR. This allows transition from double-negative to the double-positive stage of T cell development.
- Common γ-chain
-
A cytokine receptor subunit, CD132, common to receptor complexes of IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21. It is required for functional signalling downstream of these cytokines. Mutations in the common γ-chain impair lymphocyte development.
- Somatic hypermutation
-
SHM. The controlled process of mutation in the variable region of immunoglobulin locus that diversifies the range of B cell receptors used by the immune system to fine-tune the recognition of pathogens.
- E-box
-
A DNA element with a consensus sequence CANNTG that is recognized and bound by MYC and MAX transcription factors that contain the basic helix–loop–helix structural motif.
- Transcriptional pause
-
An RNA Pol II molecule that has not engaged in a productive transcription elongation and has paused at the promoter-proximal region under the control of negative regulators such as NELF and DSIF.
Rights and permissions
About this article
Cite this article
Nawijn, M., Alendar, A. & Berns, A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11, 23–34 (2011). https://doi.org/10.1038/nrc2986
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2986
This article is cited by
-
Elraglusib (formerly 9-ING-41) possesses potent anti-lymphoma properties which cannot be attributed to GSK3 inhibition
Cell Communication and Signaling (2023)
-
MicroRNAs as the pivotal regulators of cisplatin resistance in head and neck cancers
Cancer Cell International (2023)
-
The SUMOylation and ubiquitination crosstalk in cancer
Journal of Cancer Research and Clinical Oncology (2023)
-
Antitumor activity of the protein kinase inhibitor 1-(β-D-2′-deoxyribofuranosyl)-4,5,6,7-tetrabromo- 1H-benzimidazole in breast cancer cell lines
BMC Cancer (2022)
-
Exploring the mechanism and experimental verification of puerarin in the treatment of endometrial carcinoma based on network pharmacology and bioinformatics analysis
BMC Complementary Medicine and Therapies (2022)