Key Points
-
It is not only functionally related genes that cluster in the mammalian genome: there also seems to be general clustering of apparently unrelated genes.
-
Specialized genomic regions, such as the homeobox, globin and major histocompatibility complex regions, can give useful insights into how 'ordinary' regions of the genome are regulated. However, they also use specialized means of coordinate regulation that might not be generally applicable.
-
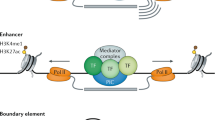
Histone modifications seem to operate at a local level in the mammalian genome — they do not generally spread over large domains.
-
Locus-control regions do not seem to operate over clusters of unrelated genes.
-
Regions of open chromatin-fibre structure are clustered in the human genome. It is suggested that this creates an environment that is permissive for gene activation by transcription factors.
-
Open chromatin regions contain clusters of broadly expressed unrelated genes. This might be a selective force for maintaining these gene clusters during evolution.
-
Gene regulation cannot be understood by considering individual genes. Whole-genome approaches, such as the use of genomic microarrays, can begin to tell us about large-scale mechanisms of gene regulation.
Abstract
Much of what we know about the chromatin-based mechanisms that regulate gene expression in mammals has come from the study of what are, paradoxically, atypical genes. These are clusters of structurally and/or functionally related genes that are coordinately regulated during development, or between different cell types. Can unravelling the mechanisms of gene regulation at these gene clusters help us to understand how other genes are controlled? Moreover, can it explain why there is clustering of apparently unrelated genes in mammalian genomes?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hurst, L. D., Pal, C. & Lercher, M. J. The evolutionary dynamics of eukaryotic gene order. Nature Rev. Genet. 5, 299–310 (2004).
De Leo, A. A. et al. Sequencing and mapping hemoglobin gene clusters in the Australian model dasyurid marsupial Sminthopsis macroura. Cytogenet. Genome Res. 108, 333–341 (2005).
Gillemans, N. et al. Functional and comparative analysis of globin loci in pufferfish and humans. Blood 101, 2842–2849 (2003).
Mason, M. M., Lee, E., Westphal, H. & Reitman, M. Expression of the chicken β-globin gene cluster in mice: correct developmental expression and distributed control. Mol. Cell. Biol. 15, 407–414 (1995).
Kmita, M. & Duboule, D. Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333 (2003).
Ikuta, T., Yoshida, N., Satoh, N. & Saiga, H. Ciona intestinalis Hox gene cluster: its dispersed structure and residual colinear expression in development. Proc. Natl Acad. Sci. USA 101, 15118–15123 (2004).
Seo, H. C. et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431, 67–71 (2004).
Patel, N. H. Evolutionary biology: time, space and genomes. Nature 431, 28–29 (2004).
Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R. & Fraser, P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5, 377–386 (2000).
Plant, K. E., Routledge, S. J. & Proudfoot, N. J. Intergenic transcription in the human β-globin gene cluster. Mol. Cell. Biol. 21, 6507–6514 (2001).
Trowsdale, J. The gentle art of gene arrangement: the meaning of gene clusters. Genome Biol. 3, COMMENT2002 (2002).
Danchin, E. G. & Pontarotti, P. Towards the reconstruction of the bilaterian ancestral pre-MHC region. Trends Genet. 20, 587–591 (2004).
Kumanovics, A., Takada, T. & Lindahl, K. F. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 21, 629–657 (2003).
Horton, R. et al. Gene map of the extended human MHC. Nature Rev. Genet. 5, 889–899 (2004).
Wang, J. et al. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164, 515–526 (2004).
Shykind, B. M. Regulation of odorant receptors: one allele at a time. Hum. Mol. Genet. 14 (Suppl. 1), R33–R39 (2005).
Amadou, C. et al. Co-duplication of olfactory receptor and MHC class I genes in the mouse major histocompatibility complex. Hum. Mol. Genet. 12, 3025–3040 (2003).
Shiina, T. et al. Genomic anatomy of a premier major histocompatibility complex paralogous region on chromosome 1q21–q22. Genome Res. 11, 789–802 (2001).
Alibardi, L. & Toni, M. Localization and characterization of specific cornification proteins in avian epidermis. Cells Tissues Organs 178, 204–215 (2004).
Williams, R. R., Broad, S., Sheer, D. & Ragoussis, J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272, 163–175 (2002).
Craig, J. M. & Bickmore, W. A. The distribution of CpG islands in mammalian chromosomes. Nature Genet. 7, 376–382 (1994).
Caron, H. et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291, 1289–1292 (2001).
Versteeg, R. et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13, 1998–2004 (2003).
Lercher, M. J., Urrutia, A. O. & Hurst, L. D. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nature Genet. 31, 180–183 (2002). This paper proposes that genes that are broadly expressed, rather than those that are highly expressed, cluster in the human genome.
Cajiao, I., Zhang, A., Yoo, E. J., Cooke, N. E. & Liebhaber, S. A. Bystander gene activation by a locus control region. EMBO J. 23, 3854–3863 (2004).
Singer, G. A., Lloyd, A. T., Huminiecki, L. B. & Wolfe, K. H. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol. Biol. Evol. 22, 767–775 (2005).
Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F. & Fraser, P. Long-range chromatin regulatory interactions in vivo. Nature Genet. 32, 1–4 (2002).
Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002). This study used the 3C (chromosome conformation capture) technique to show that regulatory and coding regions of the β-globin locus are brought into close proximity with each other.
Drissen, R. et al. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 18, 2485–2490 (2004).
Vakoc, C. R. et al. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462 (2005).
Palstra, R. J. et al. The β-globin nuclear compartment in development and erythroid differentiation. Nature Genet. 35, 190–194 (2003).
Spitz, F., Gonzalez, F. & Duboule, D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405–417 (2003).
Zakany, J., Kmita, M. & Duboule, D. A dual role for Hox genes in limb anterior–posterior asymmetry. Science 304, 1669–1672 (2004).
Carson, S. & Wiles, M. V. Far upstream regions of class II MHC Ea are necessary for position-independent, copy-dependent expression of Ea transgene. Nucleic Acids Res. 21, 2065–2072 (1993).
Krawczyk, M. et al. Long distance control of MHC class II expression by multiple distal enhancers regulated by regulatory factor X complex and CIITA. J. Immunol. 173, 6200–6210 (2004).
Forsberg, E. C. et al. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA 97, 14494–14499 (2000).
Chambeyron, S. & Bickmore, W. A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 (2004).
Rastegar, M., Kobrossy, L., Kovacs, E. N., Rambaldi, I. & Featherstone, M. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol. Cell. Biol. 24, 8090–8103 (2004).
Bernstein, B. E. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005). This large-scale analysis of histone modifications in mammalian cells reveals punctate domains of modification for most genomic regions, but broad domains of active histone modifications at Hox gene loci.
Raval, A. et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAFII250. Mol. Cell 7, 105–115 (2001).
Zika, E. & Ting, J. P. Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr. Opin. Immunol. 17, 58–64 (2005).
Masternak, K., Peyraud, N., Krawczyk, M., Barras, E. & Reith, W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nature Immunol. 4, 132–137 (2003).
Kimura, A. P., Liebhaber, S. A. & Cooke, N. E. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol. Endocrinol. 18, 1018–1032 (2004).
Roh, T. Y., Cuddapah, S. & Zhao, K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19, 542–552 (2005). This study used a novel combination of immunoprecipitation and serial analysis of gene expression (SAGE) for mapping histone modifications across the genome.
Chambeyron, S., Da Silva, N. R., Lawson, K. A. & Bickmore, W. A. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132, 2215–2223 (2005).
Ragoczy, T., Telling, A., Sawado, T., Groudine, M. & Kosak, S. T. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome. Res. 11, 513–525 (2003).
Volpi, E. V. et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113, 1565–1576 (2000). This was the first study to show that a region of the genome can be extruded from a chromosome territory in a regulated fashion.
Gilbert, N. et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibres. Cell 118, 555–566 (2004). This paper highlights the first global survey of the biophysical properties of chromatin fibres across the human genome. It also proposes that an open chromatin structure is present in regions of high gene density.
Lercher, M. J., Urrutia, A. O., Pavlicek, A. & Hurst, L. D. A unification of mosaic structures in the human genome. Hum. Mol. Genet. 12, 2411–2415 (2003).
Mahy, N. L., Perry, P. E. & Bickmore, W. A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159, 753–763 (2002).
Zhou, Y. et al. Genome-wide identification of chromosomal regions of increased tumor expression by transcriptome analysis. Cancer Res. 63, 5781–5784 (2003).
Murrell, A., Rakyan, V. K. & Beck, S. From genome to epigenome. Hum. Mol. Genet. 14 (Suppl. 1), R3–R10 (2005).
Crawford, G. E. et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl Acad. Sci. USA 101, 992–997 (2004).
Sabo, P. J. et al. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl Acad. Sci. USA 101, 4537–4542 (2004).
Weil, M. R., Widlak, P., Minna, J. D. & Garner, H. R. Global survey of chromatin accessibility using DNA microarrays. Genome Res. 14, 1374–1381 (2004).
Marenholz, I., Heizmann, C. W. & Fritz, G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 322, 1111–1122 (2004).
Marenholz, I. et al. Identification of human epidermal differentiation complex (EDC)-encoded genes by subtractive hybridization of entire YACs to a gridded keratinocyte cDNA library. Genome Res. 11, 341–355 (2001).
Marshall, D., Hardman, M. J., Nield, K. M. & Byrne, C. Differentially expressed late constituents of the epidermal cornified envelope. Proc. Natl Acad. Sci. USA 98, 13031–13036 (2001).
Huber, M. et al. Isolation and characterization of human repetin, a member of the fused gene family of the epidermal differentiation complex. J. Invest. Dermatol. 124, 998–1007 (2005).
Contzler, R., Favre, B., Huber, M. & Hohl, D. Cornulin, a new member of the 'fused gene' family, is expressed during epidermal differentiation. J. Invest. Dermatol. 124, 990–997 (2005).
Jang, S. I. & Steinert, P. M. Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1, and AP2 families. J. Biol. Chem. 277, 42268–42279 (2002).
Martin, N., Patel, S. & Segre, J. A. Long-range comparison of human and mouse Sprr loci to identify conserved noncoding sequences involved in coordinate regulation. Genome Res. 14, 2430–2438 (2004).
Johnnidis, J. B. et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc. Natl Acad. Sci. USA 102, 7233–7238 (2005).
Acknowledgements
D.S. is a Medical Research Council (MRC) pre-doctoral training fellow and W.A.B. is a Centennial fellow of the James S. McDonnell foundation. This work was funded by the MRC UK, and in part by the EU FP6 Network of Excellence Epigenome.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- IMMUNO-PROTEASOME
-
A proteasome complex that degrades proteins into peptides for presentation with MHC class I molecules.
- COLLINEARITY
-
The correspondence between the linear order of genes on the chromosome and the sequential order of their expression.
- UROCHORDATES
-
A subphylum of Chordata that are also known as tunicates. They have a notochord during their early stages of development.
- LINKAGE DISEQUILIBRIUM
-
The non-random association of alleles at adjacent loci along a chromosome.
- TELEOSTS
-
A taxonomic group that comprises most extant bony fishes.
- PML NUCLEAR BODIES
-
Sub-nuclear compartments that are defined by the presence of the PML (promyelocytic leukaemia) protein. They have been associated with diverse nuclear functions including transcription, DNA repair, viral defence, stress, cell-cycle regulation, proteolysis and apoptosis.
- LORICRIN
-
The predominant protein of the cornified envelope in keratinocytes, which is encoded by a gene in the EDC.
- FILAGGRIN
-
A protein that is involved in aggregating keratin during the terminal differentiation of epidermal keratinocytes, and is encoded by a gene in the EDC.
- SUPRABASAL LAYERS
-
Layers of progressively differentiating keratinocytes that are found above the basal layer of stem cells.
- CORNIFIED ENVELOPE
-
A tough protein–lipid structure that is formed under the plasma membrane of keratinocytes during their terminal differentiation.
- INVOLUCRIN
-
A component of the cornified envelope.
- SYNTENY
-
The preserved order of genes along a chromosome in related organisms.
- YOLK SAC
-
The first site of blood formation in the mammalian embryo.
- FLUORESCENCE IN SITU HYBRIDIZATION
-
This is a cytological technique that is used to detect and localize DNA sequences on chromosomes, or in nuclei, using fluorescent probes.
- B-LYMPHOBLASTOID CELLS
-
Peripheral blood mononuclear cells that are transformed with the Epstein–Barr virus.
- ANTIGEN-PRESENTING CELLS
-
Dendritic cells, macrophages and B cells. These cells express MHC class II genes, display foreign antigens that form complexes with MHC on their surfaces, and can activate T cells.
- SUCROSE GRADIENT SEDIMENTATION
-
An ultracentrifugation technique that separates macromolecules on the basis of their mass and their size or shape (frictional coefficient).
Rights and permissions
About this article
Cite this article
Sproul, D., Gilbert, N. & Bickmore, W. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 6, 775–781 (2005). https://doi.org/10.1038/nrg1688
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg1688
This article is cited by
-
Immune dysfunction in developmental programming of type 2 diabetes mellitus
Nature Reviews Endocrinology (2021)
-
SegCorr a statistical procedure for the detection of genomic regions of correlated expression
BMC Bioinformatics (2017)
-
Brg1-mediated Nrf2/HO-1 pathway activation alleviates hepatic ischemia–reperfusion injury
Cell Death & Disease (2017)
-
QTL analysis of cocoon shell weight identifies BmRPL18 associated with silk protein synthesis in silkworm by pooling sequencing
Scientific Reports (2017)