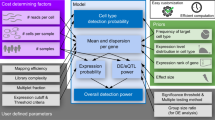
Key Points
-
We examine emerging consensus points for five key components of microarray analysis — design, preprocessing, inference, classification and validation.
-
Biological replication is important in the design of microarray experiments. The evidence indicates that a minimum of five biological cases per group should be analysed. Technical replicates are rarely warranted when testing for differential expression.
-
Modern methods (such as those used by the PowerAtlas resource) should be used for estimating the required sample size before conducting experiments.
-
mRNA pooling can be beneficial in certain cases in which identifying differential expression is the goal.
-
Microarray experiments should be designed to avoid confounding by extraneous factors.
-
Many methods exist for image processing, normalization and transformation with respect to different microarray platforms. However, which preprocessing algorithms to use and under what conditions remains an area of active research.
-
For inference in microarray experiments, test statistics for determining differential expression should consider variability; fold change does not achieve this. Test statistics that incorporate variance shrinkage are generally preferred.
-
False-discovery-rate estimation procedures are generally recommended over family-wise error rate control procedures.
-
Gene-class testing is encouraged when testing for differential expression.
-
The assessment of intersections of findings when testing multiple related propositions and how to appropriately use resampling-based inference are areas for which many questions remain.
-
Before undertaking cluster analysis, it is important to consider whether it actually addresses the question being asked and whether sufficient sample sizes can be obtained to yield reliable results.
-
When using supervised-classification procedures, cross-validation should be carried out using data that have had no role whatsoever in the derivation of the prediction rule.
-
Although discussed frequently in microarray research, validation of results is an area that requires further attention. When and how validation should be carried out, in addition to which criteria determine validation, are topics that remain to be addressed.
Abstract
In just a few years, microarrays have gone from obscurity to being almost ubiquitous in biological research. At the same time, the statistical methodology for microarray analysis has progressed from simple visual assessments of results to a weekly deluge of papers that describe purportedly novel algorithms for analysing changes in gene expression. Although the many procedures that are available might be bewildering to biologists who wish to apply them, statistical geneticists are recognizing commonalities among the different methods. Many are special cases of more general models, and points of consensus are emerging about the general approaches that warrant use and elaboration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kerr, M. K. Design considerations for efficient and effective microarray studies. Biometrics 59, 822–828 (2003).
Page, G. P., Edwards, J. W., Barnes, S., Weindruch, R. & Allison, D. B. A design and statistical perspective on microarray gene expression studies in nutrition: the need for playful creativity and scientific hard-mindedness. Nutrition 19, 997–1000 (2003).
Yang, M. C., Yang, J. J., McIndoe, R. A. & She, J. X. Microarray experimental design: power and sample size considerations. Physiol. Genomics 16, 24–28 (2003).
Kerr, M. K. & Churchill, G. A. Experimental design for gene expression microarrays. Biostatistics 2, 183–201 (2001).
Dobbin, K., Shih, J. H. & Simon, R. Statistical design of reverse dye microarrays. Bioinformatics 19, 803–810 (2003).
Churchill, G. A. Fundamentals of experimental design for cDNA microarrays. Nature Genet. 32, S490–S495 (2002).
Yang, Y. H. & Speed, T. Design issues for cDNA microarray experiments. Nature Rev. Genet. 3, 579–588 (2002).
Allison, D. B., Allison, R. L., Faith, M. S., Paultre, F. & Pi-Sunyer, F. X. Power and money: designing statistically powerful studies while minimizing financial costs. Psychol. Methods 2, 20–33 (1997).
Allison, D. B. et al. A mixture model approach for the analysis of microarray gene expression data. Comput. Stat. Data Analysis 39, 1–20 (2002). This was the first paper in the field of microarray research to introduce mixture modelling.
Pavlidis, P., Li, Q. & Noble, W. S. The effect of replication on gene expression microarray experiments. Bioinformatics 19, 1620–1627 (2003).
Tsai, C. A., Hsueh, H. M. & Chen, J. J. Estimation of false discovery rates in multiple testing: application to gene microarray data. Biometrics 59, 1071–1081 (2003).
Pan, W., Lin, J. & Le, C. T. How many replicates of arrays are required to detect gene expression changes in microarray experiments? A mixture model approach. Genome Biol. 3, research0022 (2002).
Zien, A., Fluck, J., Zimmer, R. & Lengauer, T. Microarrays: how many do you need? J. Comput. Biol. 10, 653–667 (2003).
Gadbury, G. L. et al. Power analysis and sample size estimation in the age of high dimensional biology: a parametric bootstrap approach and examples from microarray research. Stat. Methods Med. Res. 13, 325–338 (2004). This paper offers convenient FDR-based methods for power analysis and sample-size estimation in microarray and other high-dimensional testing situations.
Pawitan, Y., Michiels, S., Koscielny, S., Gusnanto, A. & Ploner, A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 21, 3017–3024 (2005).
Muller, P., Parmigiani, G., Robert, C. & Rousseau, J. Optimal sample size for multiple testing: The case of gene expression microarrays. J. Am. Stat. Assoc. 99, 990–1001 (2004).
Dobbin, K. & Simon, R. Sample size determination in microarray experiments for class comparison and prognostic classification. Biostatistics. 6, 27–38 (2005).
Garge, N., Page, G. P., Spargue, A. P., Gorman, B. S. & Allison, D. B. Reproducible clusters from microarray research: whither? BMC Bioinformatics 6 (Suppl. 2), S10 (2005). The authors evaluate clustering techniques using real data, and find that with sample sizes of less than 50, the reproducibility of results is poor.
Kendziorski, C. M., Zhang, Y., Lan, H. & Attie, A. D. The efficiency of pooling mRNA in microarray experiments. Biostatistics 4, 465–477 (2003). This paper clarifies concepts and statistical design issues that are involved with mRNA pooling in microarray experiments.
Kendziorski, C., Irizarry, R. A., Chen, K. S., Haag, J. D. & Gould, M. N. On the utility of pooling biological samples in microarray experiments. Proc. Natl Acad. Sci. USA 102, 4252–4257 (2005).
Chen, Y., Dougherty, E. R. & Bittner, M. L. Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Opt. 2, 364–374 (1997).
Schadt, E. E., Li, C., Ellis, B. & Wong, W. H. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell Biochem. Suppl. 37, 120–125 (2001).
Ekstrom, C. T., Bak, S., Kristensen, C. & Rudemo, M. Spot shape modelling and data transformations for microarrays. Bioinformatics 20, 2270–2278 (2004).
Steinfath, M. et al. Automated image analysis for array hybridization experiments. Bioinformatics 17, 634–641 (2001).
Yang, Y. H., Buckley, M. J. & Speed, T. P. Analysis of cDNA microarray images. Brief Bioinform. 2, 341–349 (2001).
Quackenbush, J. Microarray data normalization and transformation. Nature Genet. 32, 496–501 (2002).
Yang, Y. H. et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15 (2002).
Smyth, G. K. & Speed, T. Normalization of cDNA microarray data. Methods 31, 265–273 (2003).
Qin, L. X. & Kerr, K. F. Empirical evaluation of data transformations and ranking statistics for microarray analysis. Nucleic Acids Res. 32, 5471–5479 (2004). This article presents the effect of different image-processing and normalization techniques on microarray analysis conclusions.
Affymetrix. Affymetrix Expression Analysis Technical Manual (Affymetrix, Santa Clara, California, 2004).
Nielsen, H. B., Gautier, L. & Knudsen, S. Implementation of a gene expression index calculation method based on the PDNN model. Bioinformatics 21, 687–688 (2005).
Irizarry, R. A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 (2003).
Mehta, T., Tanik, M. & Allison, D. B. Towards sound epistemological foundations of statistical methods for high-dimensional biology. Nature Genet. 36, 943–947 (2004). This paper clarifies the importance of methods for evaluating the validity of proposed statistical methodologies in high-dimensional biology, with an emphasis on microarray research.
Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Choe, S. E., Boutros, M., Michelson, A. M., Church, G. M. & Halfon, M. S. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 6, R16 (2005).
Cope, L. M., Irizarry, R. A., Jaffee, H. A., Wu, Z. & Speed, T. P. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics 20, 323–331 (2004).
Chen, D. T. A graphical approach for quality control of oligonucleotide array data. J. Biopharm. Stat. 14, 591–606 (2004).
Hsiao, A., Worrall, D. S., Olefsky, J. M. & Subramaniam, S. Variance-modeled posterior inference of microarray data: detecting gene-expression changes in 3T3-L1 adipocytes. Bioinformatics 20, 3108–3127 (2004).
Miller, R. A., Galecki, A. & Shmookler-Reis, R. J. Interpretation, design, and analysis of gene array expression experiments. J. Gerontol. A 56, B52–B57 (2001).
Budhraja, V., Spitznagel, E., Schaiff, W. T. & Sadovsky, Y. Incorporation of gene-specific variability improves expression analysis using high-density DNA microarrays. BMC Biol. 1, 1 (2003).
Cui, X., Hwang, J. T., Qiu, J., Blades, N. J. & Churchill, G. A. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 6, 59–75 (2005). This article provides one method of shrinkage and compares its performance with other variance shrinkage methods.
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci USA 98, 5116–5121 (2001).
Baldi, P. & Long, A. D. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17, 509–519 (2001).
Edwards, J. W. et al. Empirical Bayes estimation of gene-specific effects in micro-array research. Funct. Integr. Genomics 5, 32–39 (2005).
Ge, Y. C., Dudoit, S. & Speed, T. P. Resampling-based multiple testing for microarray data analysis. Test 12, 1–77 (2003).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate — a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
Hsueh, H. M., Chen, J. J. & Kodell, R. L. Comparison of methods for estimating the number of true null hypotheses in multiplicity testing. J. Biopharm. Stat. 13, 675–689 (2003).
van der Lann, M. J., Dudoit, S. & Pollard, K. S. Augmentation procedures for control of the generalized family-wise error rate and tail probabilities for the proportion of false positives. Stat. Appl. Genet. Mol. Biol. 3, A15 (2004).
Storey, J. D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 (2003). This paper clarifies the key terminology and concepts used in FDR-related methods.
Do, K. A., Mueller, P. & Tang, F. A nonparametric Bayesian mixture model for gene expression. J. R. Stat. Soc. Ser. C 54, 1–18 (2005).
Pounds, S. & Morris, S. W. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 19, 1236–1242 (2003).
Datta, S. & Datta, S. Empirical Bayes screening of many p-values with applications to microarray studies. Bioinformatics 21, 1987–1994 (2005).
Efron, B., Tibshirani, R., Storey, J. D. & Tusher, V. G. Empirical Bayes analysis of a microarray experiment. J. Am. Stat. Assoc. 96, 1151–1160 (2001).
Newton, M. A., Noueiry, A., Sarkar, D. & Ahlquist, P. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics 5, 155–176 (2004).
Newton, M. A., Kendziorski, C. M., Richmond, C. S., Blattner, F. R. & Tsui, K. W. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J. Comput. Biol. 8, 37–52 (2001).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 34, 267–273 (2003).
Osier, M. V. in DNA Microarrays and Statistical Genomic Techniques: Design, Analysis, and Interpretation of Experiments (Marcel Dekker, New York, 2005).
Osier, M. V., Zhao, H. & Cheung, K. H. Handling multiple testing while interpreting microarrays with the Gene Ontology Database. BMC Bioinformatics 5, 124 (2004).
Khatri, P., Draghici, S., Ostermeier, G. C. & Krawetz, S. A. Profiling gene expression using onto-express. Genomics 79, 266–270 (2002).
Pavlidis, P., Weston, J., Cai, J. & Noble, W. S. Learning gene functional classifications from multiple data types. J. Comput. Biol. 9, 401–411 (2002).
Pavlidis, P., Qin, J., Arango, V., Mann, J. J. & Sibille, E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem. Res. 29, 1213–1222 (2004). This study introduces a gene-class testing method that uses the full continuous evidence that is available within p -values.
Ben Shaul, Y., Bergman, H. & Soreq, H. Identifying subtle interrelated changes in functional gene categories using continuous measures of gene expression. Bioinformatics 21, 1129–1137 (2005).
Zeeberg, B. R. et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4, R28 (2003).
Damian, D. & Gorfine, M. Statistical concerns about the GSEA procedure. Nature Genet. 36, 663 (2004).
Persson, S., Wei, H., Milne, J., Page, G. P. & Somerville, C. R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl Acad. Sci. USA 102, 8633–8638 (2005).
Kyng, K. J., May, A., Kolvraa, S. & Bohr, V. A. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc. Natl Acad. Sci. USA 100, 12259–12264 (2003).
Schmid, C. H., Lau, J., McIntosh, M. W. & Cappelleri, J. C. An empirical study of the effect of the control rate as a predictor of treatment efficacy in meta-analysis of clinical trials. Stat. Med. 17, 1923–1942 (1998).
Berger, R. L. Multiparameter hypothesis testing and acceptance sampling. Technometrics 24, 295–300 (1982).
Neuhauser, M., Boes, T. & Jockel, K. H. Two-part permutation tests for DNA methylation and microarray data. BMC Bioinformatics 6, 35 (2005).
Barry, W. T., Nobel, A. B. & Wright, F. A. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics 21, 1943–1949 (2005).
Pan, W. On the use of permutation in and the performance of a class of nonparametric methods to detect differential gene expression. Bioinformatics 19, 1333–1340 (2003).
Xu, R. H. & Li, X. C. A comparison of parametric versus permutation methods with applications to general and temporal microarray gene expression data. Bioinformatics 19, 1284–1289 (2003).
Landgrebe, J., Wurst, W. & Welzl, G. Permutation-validated principal components analysis of microarray data. Genome Biol. 3, RESEARCH0019 (2002).
Troendle, J. F., Korn, E. L. & McShame, L. M. An example of slow convergence of the bootstrap in high dimensions. Am. Stat. 58, 25–29 (2004). This presents an excellent overview of the nuances of resampling methodology that is used in microarray research, and discusses the fact that such methods are not assumption-free panaceas that are valid under all circumstances.
Kennedy, P. E. & Cade, B. S. Randomization tests for multiple regression. Commun. Stat. 25, 923–936 (1996).
Gadbury, G. L., Page, G. P., Heo, M., Mountz, J. D. & Allison, D. B. Randomization tests for small samples: an application for genetic expression data. J. R. Stat. Soc. Ser. C 52, 365–376 (2003).
Yeung, K. Y., Haynor, D. R. & Ruzzo, W. L. Validating clustering for gene expression data. Bioinformatics 17, 309–318 (2001).
Datta, S. & Datta, S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics 19, 459–466 (2003).
Shih, J. H. et al. Effects of pooling mRNA in microarray class comparisons. Bioinformatics 20, 3318–3325 (2004).
Yeung, K. Y., Medvedovic, M. & Bumgarner, R. E. From co-expression to co-regulation: how many microarray experiments do we need? Genome Biol. 5, R48 (2004).
Bryan, J. Problems in gene clustering based on gene expression data. J. Multivariate Analysis 90, 44–66 (2004). This is an excellent overview of the methodological and conceptual challenges in the use of cluster analysis in gene-expression studies.
Kerr, M. K. & Churchill, G. A. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc. Natl Acad. Sci. USA 98, 8961–8965 (2001).
Zhang, K. & Zhao, H. Assessing reliability of gene clusters from gene expression data. Funct. Integr. Genomics 1, 156–173 (2000).
Tseng, G. C. & Wong, W. H. Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics 61, 10–16 (2005).
Horth, J. Computer Intensive Statistical Methods Validation, Model Selection and Boostrap (Chapman and Hall, London, 1994).
Ambroise, C. & McLachlan, G. J. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl Acad. Sci. USA 99, 6562–6566 (2002). This article addresses selection bias in the context of predictive error-estimation and cross-validation for microarray studies.
Furlanello, C., Serafini, M., Merler, S. & Jurman, G. Entropy-based gene ranking without selection bias for the predictive classification of microarray data. BMC Bioinformatics 4, 54 (2003).
Fu, W. J., Carroll, R. J. & Wang, S. Estimating misclassification error with small samples via bootstrap cross-validation. Bioinformatics 21, 1979–1986 (2005).
Dobbin, K. & Simon, R. Sample size determination in microarray experiments for class comparison and prognostic classification. Biostatistics 6, 27–38 (2005).
Hwang, D., Schmitt, W. A., Stephanopoulos, G. & Stephanopoulos, G. Determination of minimum sample size and discriminatory expression patterns in microarray data. Bioinformatics 18, 1184–1193 (2002).
Mukherjee, S. et al. Estimating dataset size requirements for classifying DNA microarray data. J. Comput. Biol. 10, 119–142 (2003).
Rajeevan, M. S., Ranamukhaarachchi, D. G., Vernon, S. D. & Unger, E. R. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25, 443–451 (2001).
Rockett, J. C. & Hellmann, G. M. Confirming microarray data — is it really necessary? Genomics 83, 541–549 (2004).
Rocke, D. M. & Durbin, B. Approximate variance-stabilizing transformations for gene-expression microarray data. Bioinformatics 19, 966–972 (2003).
Pounds, S. & Cheng, C. Statistical development and evaluation of microarray gene expression data filters. J. Comput. Biol. 12, 482–495 (2005).
Acknowledgements
The authors are supported in part by grants from the US National Institutes of Health, National Science Foundation and Department of Defense. We are grateful to C. Kendziorski for her helpful discussion on this document.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
OMIM
FURTHER INFORMATION
A free online microarray analysis course from the University of Alabama at Birmingham
ArrayExpress microarray data repository
BioConductor open source software for bioinformatics
ermineJ — Gene Ontology analysis for microarry data
Gene Expression Omnibus data repository
HDBStat! High Dimension Biology Statistical analysis software
Glossary
- Fold change
-
A metric for comparing a gene's mRNA-expression level between two distinct experimental conditions. Its arithmetic definition differs between investigators.
- Case
-
In a microarray experiment, a case is the biological unit under study; for example, one soybean, one mouse or one human.
- Power
-
This is classically defined as the probability of rejecting a null hypothesis that is false. However, power has been defined in several ways for microarray studies.
- False-discovery rate
-
(FDR). The expected proportion of rejected null hypotheses that are false positives. When no null hypotheses are rejected, FDR is taken to be zero.
- Normalization
-
The process by which microarray spot intensities are adjusted to take into account the variability across different experiments and platforms.
- Transformation
-
The application of a specific mathematical function so that data are changed into a different form. Often, the new form of the data satisfies assumptions of statistical tests. The most common transformation in microarray studies is log2.
- Plasmode
-
A real (not computer simulated) data set for which the true structure is known and is used as a way of testing a proposed analytical method.
- Parameter
-
A quantity (for example, mean) that characterizes some aspect of a (usually theoretically infinite) population.
- Type 1 error
-
A false positive, or the rejection of a true null hypothesis; for example, declaring a gene to be differentially expressed when it is not.
- Type 2 error
-
A false negative, or failing to reject a false null hypothesis; for example, not declaring a gene to be differentially expressed when it is.
- Long-range error rate
-
The expected error rate if experiments and analyses of the type under consideration were repeated an infinite number of times.
- t-tests
-
Statistical tests that are used to determine a statistically significant difference between two groups by looking at differences between two independent means.
- ANOVA
-
Analysis of variance. A statistical test for determining differences in mean values between two or more groups.
- Logistic regression
-
A regression technique that is used in cases where the outcome variable is binary (dichotomous).
- Survival analysis
-
A statistical methodology for analysing time-to-event data.
- α-value
-
The nominal probability (set by the investigator) of making a type 1 error.
- Bonferroni correction
-
A family-wise error rate (FWER) control procedure that sets the α-value level for each test and strongly controls the FWER for any dependency structure among the tests.
- Bayesian probability
-
The probability of a proposition being true, which is conditional on the observed data.
- Gene Ontology
-
A way of describing gene products in terms of their associated biological processes, cellular components and molecular functions in a species-independent manner.
- Null hypothesis
-
The hypothesis that is being tested in a statistical test. Typically in a microarray setting it is the hypothesis that states: there is no difference between gene-expression levels across groups or conditions.
- p-value
-
The probability, were the null hypothesis true, of obtaining results that are as discrepant or more discrepant from those expected under the null hypothesis than those actually obtained.
- Permutation test
-
A statistical hypothesis test in which some elements of the data are permuted (shuffled) to create multiple new pseudo-data sets. One then evaluates whether a statistic quantifying departure from the null hypothesis is greater in the observed data than a large proportion of the corresponding statistics calculated on the multiple pseudo-data sets.
- Intersection-union tests
-
Multicomponent tests in which the compound null hypothesis consists of the union of two or more component null hypotheses.
- Chi-square test of independence
-
A test of the independence of two categorical variables that is based on the chi-square distribution. The test is valid only under the assumption that all cases are independent.
- Min-test
-
A statistical IUT test in which the union of a null hypotheses is rejected if, and only if, for each component null hypothesis the p-value <α.
- Posterior probability
-
The Bayesian probability that a hypothesis is correct, which is conditional on the observed data.
- Bootstrap analysis
-
A form of computer-intensive resampling-based inference. Pseudo-data sets are created by sampling from the observed data with replacement (that is, after a case is resampled, it is returned to the original data and can, potentially, be drawn again).
- Sampling variation
-
The variability in statistics that occurs among random samples from the same population and is due solely to the process of random sampling.
- Overfitting
-
This occurs when an excessively complex model with too many parameters is developed from a small sample of 'training' data. The model fits those data well, but does so by capitalizing on chance variations and, therefore, will fit a fresh set 'test' data poorly.
- Selection bias
-
This occurs when the prediction accuracy of a rule is estimated using cases that had some role in the derivation of the rule. It is an upward bias — that is, one that overestimates the predictive accuracy.
- Operational validation
-
Re-testing a hypothesis using the original methodology (also referred to as operational replication).
- Constructive validation
-
Testing a hypothesis through a different methodology (also referred to as constructive replication).
Rights and permissions
About this article
Cite this article
Allison, D., Cui, X., Page, G. et al. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7, 55–65 (2006). https://doi.org/10.1038/nrg1749
Issue Date:
DOI: https://doi.org/10.1038/nrg1749
This article is cited by
-
The amniotic fluid proteome changes with gestational age in normal pregnancy: a cross-sectional study
Scientific Reports (2022)
-
Differential gene expression in iPSC-derived human intestinal epithelial cell layers following exposure to two concentrations of butyrate, propionate and acetate
Scientific Reports (2022)
-
Small genetic variation affecting mRNA isoforms associated with marbling and meat color in beef cattle
Functional & Integrative Genomics (2022)
-
Identification of potential hub genes associated with skin wound healing based on time course bioinformatic analyses
BMC Surgery (2021)
-
Transcriptome comparisons of in vitro intestinal epithelia grown under static and microfluidic gut-on-chip conditions with in vivo human epithelia
Scientific Reports (2021)