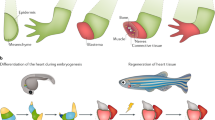
Key Points
-
Regeneration is widely distributed among the various phyla that compose the animal kingdom, including vertebrates.
-
Limitations in the ability to interrogate this attribute at the molecular level have hampered efforts to delineate the mechanistic underpinnings of regeneration.
-
Loss-of-function screens (using RNAi) and gain-of-function assays (transgenesis) have recently been introduced to study molecular pathways in traditional model systems of regeneration, overcoming past limitations to probe their biology at the molecular level.
-
Studies in simple animals such as hydra and planarians are beginning to contribute to our understanding of tissue remodelling and adult somatic stem-cell regulation in animals.
-
Studies in mammals and other vertebrates have highlighted central roles for the activation of specific signalling pathways in the processes of organ and limb regeneration.
-
Comparative studies of regenerative processes among the various animals that are currently under investigation could provide important insights into the permissive and non-permissive mechanisms that underlie regenerative competence.
-
Such information is likely to yield fundamental insights for our understanding of metazoan biology, and to expand the repertoire of therapeutic possibilities in the fields of regenerative medicine.
Abstract
Significant progress has recently been made in our understanding of animal regenerative biology, spurred on by the use of a wider range of model organisms and an increasing ability to use genetic tools in traditional models of regeneration. This progress has begun to delineate differences and similarities in the regenerative capabilities and mechanisms among diverse animal species, and to address some of the key questions about the molecular and cell biology of regeneration. Our expanding knowledge in these areas not only provides insights into animal biology in general, but also has important implications for regenerative medicine and stem-cell biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sánchez Alvarado, A. in Keywords and Concepts in Evolutionary Developmental Biology (eds Hall, B. K. & Olson, W. M.) (Harvard University Press, Cambridge, 2003).
Sánchez Alvarado, A. Regeneration in the metazoans: why does it happen? Bioessays 22, 578–590 (2000).
Lenhoff, S. G. & Lenhoff, H. M. Hydra and the Birth of Experimental Biology, 1744: Abraham Trembley's Memoirs Concerning the Natural History of a Type of Freshwater Polyp with Arms Shaped Like Horns (Boxwood Press, Pacific Grove, 1986).
Brusca, R. C. & Brusca, G. J. Invertebrates (Sinauer Associates, Sunderland, 1990).
Galliot, B. Signaling molecules in regenerating hydra. Bioessays 19, 37–46 (1997).
Martinez, D. E. Mortality patterns suggest lack of senescence in hydra. Exp. Gerontol. 33, 217–225 (1998).
Holstein, T. W., Hobmayer, E. & David, C. N. Pattern of epithelial cell cycling in hydra. Dev. Biol. 148, 602–611 (1991).
Wolpert, L., Hicklin, J. & Hornbruch, A. Positional information and pattern regulation in regeneration of hydra. Symp. Soc. Exp. Biol. 25, 391–415 (1971).
Adler, G., Hupp, T. & Kern, H. F. Course and spontaneous regression of acute pancreatitis in the rat. Virchows Archiv. 382, 31–47 (1979).
Hao, E. et al. β-cell differentiation from nonendocrine epithelial cells of the adult human pancreas. Nature Med. 12, 310–316 (2006).
Hobmayer, B. et al. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407, 186–189 (2000). Demonstrates the ancient conservation of the signalling pathways that are involved in axial specification in simple multicellular organisms.
Meinhardt, H. The radial-symmetric hydra and the evolution of the bilateral body plan: an old body became a young brain. Bioessays 24, 185–191 (2002).
Gauchat, D. et al. The orphan COUP-TF nuclear receptors are markers for neurogenesis from cnidarians to vertebrates. Dev. Biol. 275, 104–123 (2004).
Gauchat, D., Kreger, S., Holstein, T. & Galliot, B. prdl-a, a gene marker for hydra apical differentiation related to triploblastic paired-like head-specific genes. Development 125, 1637–1645 (1998).
Schummer, M., Scheurlen, I., Schaller, C. & Galliot, B. HOM/HOX homeobox genes are present in hydra (Chlorohydra viridissima) and are differentially expressed during regeneration. EMBO J. 11, 1815–1823 (1992).
Technau, U. & Bode, H. R. HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development 126, 999–1010 (1999).
Broun, M., Sokol, S. & Bode, H. R. Cngsc, a homologue of goosecoid, participates in the patterning of the head, and is expressed in the organizer region of Hydra. Development 126, 5245–5254 (1999).
Guder, C. et al. An ancient Wnt-Dickkopf antagonism in Hydra. Development 133, 901–911 (2006).
Sudhop, S. et al. Signalling by the FGFR-like tyrosine kinase, Kringelchen, is essential for bud detachment in Hydra vulgaris. Development 131, 4001–4011 (2004).
Leontovich, A. A., Zhang, J., Shimokawa, K., Nagase, H. & Sarras, M. P. Jr. A novel hydra matrix metalloproteinase (HMMP) functions in extracellular matrix degradation, morphogenesis and the maintenance of differentiated cells in the foot process. Development 127, 907–920 (2000).
Miljkovic, M., Mazet, F. & Galliot, B. Cnidarian and bilaterian promoters can direct GFP expression in transfected hydra. Dev. Biol. 246, 377–390 (2002).
Wittlieb, J., Khalturin, K., Lohmann, J. U., Anton-Erxleben, F. & Bosch, T. C. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl Acad. Sci. USA 103, 6208–6211 (2006).
Chera, S. et al. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J. Cell Sci. 119, 846–857 (2006). The first report of a phenotype that was produced by RNAi feeding in hydra and mimics a human condition.
Reddien, P. W. & Sánchez Alvarado, A. Fundamentals of Planarian Regeneration. Annu. Rev. Cell Dev. Biol. 20, 725–757 (2004).
Newmark, P. A. & Sanchez Alvarado, A. Not your father's planarian: a classic model enters the era of functional genomics. Nature Rev. Genet. 3, 210–219 (2002).
Umesono, Y., Watanabe, K. & Agata, K. A planarian orthopedic homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev. Growth Differ. 39, 723–727 (1997).
Mineta, K. et al. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc. Natl Acad. Sci. USA 100, 7666–7671 (2003).
Nakazawa, M. et al. Search for the evolutionary origin of a brain: planarian brain characterized by microarray. Mol. Biol. Evol. 20, 784–791 (2003).
Orii, H. et al. The planarian HOM/HOX homeobox genes (Plox) expressed along the anteroposterior axis. Dev. Biol. 210, 456–468 (1999).
Sánchez Alvarado, A., Newmark, P. A., Robb, S. M. & Juste, R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659–5665 (2002).
Zayas, R. M. et al. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl Acad. Sci. USA 102, 18491–18496 (2005).
Sánchez Alvarado, A. & Newmark, P. A. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl Acad. Sci. USA 96, 5049–5054 (1999).
Cebria, F. et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620–624 (2002).
Reddien, P. W., Bermange, A. L., Murfitt, K. J., Jennings, J. R. & Sánchez Alvarado, A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635–649 (2005). The first systematic RNAi screen for regeneration-specific genes in animals.
Reddien, P. W., Oviedo, N. J., Jennings, J. R., Jenkin, J. C. & Sánchez Alvarado, A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330 (2005). The first demonstration that an argonaute/PIWI molecule that is expressed in stem cells can regulate the differentiation of their division progeny.
Deng, W. & Lin, H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 (2002).
Kuramochi-Miyagawa, S. et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839–849 (2004).
French, V. Leg regeneration in the cockroach, Blatella germanica. II. Regeneration from a non-congruent tibial graft/host junction. J. Embryol. Exp. Morphol. 35, 267–301 (1976).
Uetz, G. W., McClintock, W. J., Miller, D., Smith, E. I. & Cook, K. K. Limb regeneration and subsequent asymmetry in a male secondary sexual character influences sexual selection in wolf spiders. Behav. Ecol. Sociobiol. 38, 253–257 (1996).
Haupt, J. & Coineau, Y. Moulting and morphogenesis of sensilla in a prostigmate mite (Acari, Actinotrichida, Actinedida: Caeculidae). Cell Tissue Res. 186, 63–79 (1978).
Sustar, A. & Schubiger, G. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell 120, 383–393 (2005).
Bryant, P. J. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Dev. Biol. 26, 637–651 (1971).
Monika, C. M. & Müller, L. H. Ground plan of the polychaete brain — I. Patterns of nerve development during regeneration in Dorvillea bermudensis (Dorvilleidae). J. Comp. Neurol. 471, 49–58 (2004).
Paulus, T. & Müller, M. C. Cell proliferation dynamics and morphological differentiation during regeneration in Dorvillea bermudensis (Polychaeta, Dorvilleidae). J. Morphol. 267, 393–403 (2006).
Myohara, M., Yoshida-Noro, C., Kobari, F. & Tochinai, S. Fragmenting oligochaete Enchytraeus japonensis: a new material for regeneration study. Dev. Growth Differ. 41, 549–555 (1999).
Bely, A. E. & Wray, G. A. Evolution of regeneration and fission in annelids: insights from engrailed- and orthodenticle-class gene expression. Development 128, 2781–2791 (2001).
Garcia-Arraras, J. E. et al. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea:Echinodermata). J. Exp. Zool. 281, 288–304 (1998).
Whittaker, J. R. Siphon regeneration in Ciona. Nature 255, 224–225 (1975).
Lauzon, R. J., Ishizuka, K. J. & Weissman, I. L. Cyclical generation and degeneration of organs in a colonial urochordate involves crosstalk between old and new: a model for development and regeneration. Dev. Biol. 249, 333–348 (2002).
Cox, D. N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998).
Harrisingh, M. C. et al. The Ras/Raf/ERK signalling pathway drives Schwann cell dedifferentiation. EMBO J. 23, 3061–3071 (2004).
Morrison, J. I., Loof, S., He, P. & Simon, A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J. Cell Biol. 172, 433–440 (2006).
Tsonis, P. A. Regeneration in vertebrates. Dev. Biol. 221, 273–284 (2000).
Habermann, B. et al. An Ambystoma mexicanum EST sequencing project: analysis of 17, 352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biol. 5, R67 (2004).
Putta, S. et al. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genom. 5, 54 (2004).
Sobkow, L., Epperlein, H. H., Herklotz, S., Straube, W. L. & Tanaka, E. M. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev. Biol. 290, 386–397 (2006). Describes an important experimental tool for understanding cellular contributions during regeneration.
Amaya, E. Xenomics. Genome Res. 15, 1683–1691 (2005).
Imokawa, Y. & Brockes, J. P. Selective activation of thrombin is a critical determinant for vertebrate lens regeneration. Curr. Biol. 13, 877–881 (2003).
Tanaka, E. M., Drechsel, D. N. & Brockes, J. P. Thrombin regulates S-phase re-entry by cultured newt myotubes. Curr. Biol. 9, 792–799 (1999).
Kumar, A., Velloso, C. P., Imokawa, Y. & Brockes, J. P. The regenerative plasticity of isolated urodele myofibers and its dependence on MSX1. PLoS Biol. 2, e218 (2004). Demonstrates that the requisite cellularization that is observed after the dedifferentiation of myofibres involves msx1.
Cannata, S. M., Bagni, C., Bernardini, S., Christen, B. & Filoni, S. Nerve-independence of limb regeneration in larval Xenopus laevis is correlated to the level of fgf2 mRNA expression in limb tissues. Dev. Biol. 231, 436–446 (2001).
Brockes, J. P. & Kintner, C. R. Glial growth factor and nerve-dependent proliferation in the regeneration blastema of Urodele amphibians. Cell 45, 301–306 (1986).
Kiffmeyer, W. R., Tomusk, E. V. & Mescher, A. L. Axonal transport and release of transferrin in nerves of regenerating amphibian limbs. Dev. Biol. 147, 392–402 (1991).
Yntema, C. L. Regeneration in sparsely innervated and aneurogenic forelimbs of Amblystoma larvae. J. Exp. Zool. 140, 101–123 (1959).
Irvin, B. C. & Tassava, R. A. Effects of peripheral nerve implants on the regeneration of partially and fully innervated urodele forelimbs. Wound Repair Regen. 6, 382–387 (1998).
Tsonis, P. A. Limb Regeneration (Cambridge Univ. Press, Cambridge, 1996).
Poulin, M. L., Patrie, K. M., Botelho, M. J., Tassava, R. A. & Chiu, I. M. Heterogeneity in the expression of fibroblast growth factor receptors during limb regeneration in newts (Notophthalmus viridescens). Development 119, 353–361 (1993).
Yokoyama, H. et al. Mesenchyme with fgf-10 expression is responsible for regenerative capacity in Xenopus limb buds. Dev. Biol. 219, 18–29 (2000).
Yokoyama, H., Ide, H. & Tamura, K. FGF-10 stimulates limb regeneration ability in Xenopus laevis. Dev. Biol. 233, 72–79 (2001).
Mullen, L. M., Bryant, S. V., Torok, M. A., Blumberg, B. & Gardiner, D. M. Nerve dependency of regeneration: the role of Distal-less and FGF signaling in amphibian limb regeneration. Development 122, 3487–3497 (1996).
Goss, R. J. Principles of Regeneration (Academic, New York, 1969).
Echeverri, K. & Tanaka, E. M. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science 298, 1993–1996 (2002).
Beck, C. W., Christen, B. & Slack, J. M. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell 5, 429–439 (2003). Describes the role of BMP, Msx1 and Notch in the induction of tail regeneration in frogs.
Del Rio-Tsonis, K. & Tsonis, P. A. Eye regeneration at the molecular age. Dev. Dyn. 226, 211–224 (2003).
Grogg, M. W. et al. BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration. Nature 438, 858–862 (2005). The first report to pinpoint possible regulators (BMP and Six3) of lens regeneration in the newt.
Stierwald, M., Yanze, N., Bamert, R. P., Kammermeier, L. & Schmid, V. The Sine oculis/Six class family of homeobox genes in jellyfish with and without eyes: development and eye regeneration. Dev. Biol. 274, 70–81 (2004).
Zuber, M. E., Gestri, G., Viczian, A. S., Barsacchi, G. & Harris, W. A. Specification of the vertebrate eye by a network of eye field transcription factors. Development 130, 5155–5167 (2003). An important paper that delineates the molecular pathways that are involved in eye development.
Bader, D. & Oberpriller, J. O. Repair and reorganization of minced cardiac muscle in the adult newt (Notophthalmus viridescens). J. Morphol. 155, 349–357 (1978).
Taylor, R. R. & Forge, A. Hair cell regeneration in sensory epithelia from the inner ear of a urodele amphibian. J. Comp. Neurol. 484, 105–120 (2005).
Schnapp, E. & Tanaka, E. M. Quantitative evaluation of morpholino-mediated protein knockdown of GFP, MSX1, and PAX7 during tail regeneration in Ambystoma mexicanum. Dev. Dyn. 232, 162–170 (2005).
Woods, I. G. et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 15, 1307–1314 (2005).
Peterson, R. T., Link, B. A., Dowling, J. E. & Schreiber, S. L. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl Acad. Sci. USA 97, 12965–12969 (2000).
White, J. A., Boffa, M. B., Jones, B. & Petkovich, M. A zebrafish retinoic acid receptor expressed in the regenerating caudal fin. Development 120, 1861–1872 (1994).
Nechiporuk, A. & Keating, M. T. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development 129, 2607–2617 (2002).
Poss, K. D. et al. Roles for Fgf signaling during zebrafish fin regeneration. Dev. Biol. 222, 347–358 (2000).
Thummel, R. et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev. Dyn. 235, 336–346 (2006).
Whitehead, G. G., Makino, S., Lien, C. L. & Keating, M. T. fgf20 is essential for initiating zebrafish fin regeneration. Science 310, 1957–1960 (2005). The first paper to identify a mutation in fgf20 that is linked to the inhibition of fin regeneration.
Del Rio-Tsonis, K., Washabaugh, C. H. & Tsonis, P. A. The mutant axolotl Short toes exhibits impaired limb regeneration and abnormal basement membrane formation. Proc. Natl Acad. Sci. USA 89, 5502–5506 (1992).
Makino, S. et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc. Natl Acad. Sci. USA 102, 14599–14604 (2005).
Haynes, T. & Del Rio-Tsonis, K. Retina repair, stem cells and beyond. Curr. Neurovasc. Res. 1, 231–239 (2004).
Oliver, G., Loosli, F., Koster, R., Wittbrodt, J. & Gruss, P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech. Dev. 60, 233–239 (1996).
Poss, K. D., Wilson, L. G. & Keating, M. T. Heart regeneration in zebrafish. Science 298, 2188–2190 (2002).
Raya, A. et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl Acad. Sci. USA 100, 11889–11895 (2003).
Han, M., Yang, X., Farrington, J. E. & Muneoka, K. Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development 130, 5123–5132 (2003).
Odelberg, S. J., Kollhoff, A. & Keating, M. T. Dedifferentiation of mammalian myotubes induced by msx1. Cell 103, 1099–1109 (2000).
Lesurtel, M. et al. Platelet-derived serotonin mediates liver regeneration. Science 312, 104–107 (2006).
Mastellos, D., Papadimitriou, J. C., Franchini, S., Tsonis, P. A. & Lambris, J. D. A novel role of complement: mice deficient in the fifth component of complement (C5) exhibit impaired liver regeneration. J. Immunol. 166, 2479–2486 (2001). Describes an unsuspected new role of complement during liver regeneration in mice that lack C5.
Michalopoulos, G. K. & DeFrances, M. Liver regeneration. Adv. Biochem. Eng. Biotechnol. 93, 101–134 (2005).
Susick, R. et al. Hepatic progenitors and strategies for liver cell therapies. Ann. NY Acad. Sci. 944, 398–419 (2001).
Vessey, C. J. & de la Hall, P. M. Hepatic stem cells: a review. Pathology 33, 130–141 (2001).
Gritti, A. et al. Multipotent neural stem cells reside into the rostral extension and olfactory bulb of adult rodents. J. Neurosci. 22, 437–445 (2002).
Buchli, A. D. & Schwab, M. E. Inhibition of Nogo: a key strategy to increase regeneration, plasticity and functional recovery of the lesioned central nervous system. Ann. Med. 37, 556–567 (2005).
Klimaschewski, L., Nindl, W., Feurle, J., Kavakebi, P. & Kostron, H. Basic fibroblast growth factor isoforms promote axonal elongation and branching of adult sensory neurons in vitro. Neuroscience 126, 347–353 (2004).
Condic, M. L. Adult neuronal regeneration induced by transgenic integrin expression. J. Neurosci. 21, 4782–4788 (2001).
Murry, C. E. et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature 428, 664–668 (2004).
Carroll, S. B., Grenier, J. K. & Weatherbee, S. D. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design (Blackwell Science, Malden, 2001).
Adoutte, A. et al. The new animal phylogeny: reliability and implications. Proc. Natl Acad. Sci. USA 97, 4453–4456 (2000).
Miller, D. J. & Ball, E. E. Animal evolution: the enigmatic phylum placozoa revisited. Curr. Biol. 15, R26–R28 (2005).
Delsuc, F., Brinkmann, H., Chourrout, D. & Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006).
Acknowledgements
A.S.A. thanks the National Institutes of Health and the National Institute of General Medical Sciences for supporting work on planarian regeneration, and the National Human Genome Research Institute for supporting the sequencing of the S. mediterranea genome. P.A.T. would like to thank the National Institutes of Health and the National Eye Institute for supporting eye regeneration research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Dedifferentiation
-
The process by which a terminally differentiated cell loses its tissue-specific characteristics and becomes undifferentiated. Dedifferentiated cells can either re-differentiate into cells of their original type or to a cell of different lineage.
- Transdifferentiation
-
The process by which a terminally differentiated cell dedifferentiates and then re-differentiates to a cell of a different lineage, for example, the transdifferentiation of iris pigment epithelial cells to lens during newt lens regeneration.
- Diploblast
-
An organism that is derived from two primary germ layers: the ectoderm and the endoderm.
- Triploblast
-
An organism that is derived from three primary germ layers: the ectoderm, the mesoderm and the endoderm.
- Organizer
-
The regions within an embryo that control development and differentiation.
- Autophagy
-
A nutritionally and developmentally regulated process that is involved in the intracellular destruction of endogenous proteins and the removal of damaged organelles.
- Mixoploid
-
An organism that contains cells which are of different ploidy, for example, diploid and polyploid.
- Argonaute/PIWI family
-
Members of this protein family contain PAZ and PIWI domains, which are involved, respectively, in binding small RNAs and mediating silencing, either by cleavage of mRNAs or through inhibition of translation.
- Schwann cells
-
Non-neuronal cells that mainly provide myelin insulation to axons in the peripheral nervous system of jawed vertebrates.
- Neotenous animals
-
Animals that, as adults, retain traits that are usually seen only in juveniles.
- Morpholino
-
A chemically modified oligonucleotide that behaves as an antisense RNA analogue and can therefore be used to interfere with gene function.
- Forebrain
-
The rostral-most portion of the brain.
- Complement system
-
A biochemical cascade that is involved in innate immunity: the first line of defence that helps to clear pathogens from an organism.
- Hippocampus
-
A part of the brain that is located inside the temporal lobe. It forms part of the limbic system and has a role in memory and spatial navigation.
Rights and permissions
About this article
Cite this article
Alvarado, A., Tsonis, P. Bridging the regeneration gap: genetic insights from diverse animal models. Nat Rev Genet 7, 873–884 (2006). https://doi.org/10.1038/nrg1923
Issue Date:
DOI: https://doi.org/10.1038/nrg1923
This article is cited by
-
Regulation of chromatin organization during animal regeneration
Cell Regeneration (2023)
-
Enduring questions in regenerative biology and the search for answers
Communications Biology (2023)
-
Regeneration of starfish radial nerve cord restores animal mobility and unveils a new coelomocyte population
Cell and Tissue Research (2023)
-
Molecular machineries of ciliogenesis, cell survival, and vasculogenesis are differentially expressed during regeneration in explants of the demosponge Halichondria panicea
BMC Genomics (2022)
-
Culturomics revealed the bacterial constituents of the microbiota of a 10-year-old laboratory culture of planarian species S. mediterranea
Scientific Reports (2021)