Key Points
-
The endoplasmic reticulum (ER) is a crucial intracellular organelle that is involved in the biosynthesis of cellular molecules, including proteins, carbohydrates and lipids. These become constituents of the ER or are transported to the Golgi, plasma membrane, endosomes and lysosomes.
-
The ER is involved in the transport of both MHC class I and class II complexes to the cell surface. It is the site of peptide loading onto MHC class I molecules, and it is important for regulation of the secretion of soluble immune mediators.
-
Perturbation in ER–Golgi biogenesis affects antigen presentation and immune effector functions, so it is not surprising that the ER is a target for some pathogens.
-
Several pathogens have developed strategies to modulate phagosome maturation and therefore avoid phagosome–lysosome fusion and the subsequent degradation of the pathogen: for example, by generating an ER-derived vacuole. In some cases, this has the added benefit of disrupting ER function.
-
Toxoplasma gondii, Brucella spp. and Legionella pneumophila depend on ER-derived niches for growth, but the precise effects of these pathogen–ER interactions on the immune response are still unclear.
-
Several viruses target the ER and interfere with antigen presentation by MHC class I molecules: for example, hepatitis C virus and human cytomegalovirus.
-
Despite this, the ER contributes to host defence against microorganisms, through its role in autophagy, as well as through the ER stress response and the interferon-induced ER-resident proteins (such as viperin and the p47 GTPases).
Abstract
A key determinant for the survival of intracellular pathogens is their ability to subvert the cellular processes of the host to establish a compartment that allows replication. Although most microorganisms internalized by host cells are efficiently cleared following fusion with lysosomes, many pathogens have evolved mechanisms to escape this degradation. In this Review, we provide insight into the molecular processes that are targeted by pathogens that interact with the endoplasmic reticulum and thereby subvert the immune response, ensure their survival intracellularly and cause disease. We also discuss how the endoplasmic reticulum 'strikes back' and controls microbial growth.
Similar content being viewed by others
Main
Different pathogens have evolved distinct strategies to control their intracellular fate and promote their survival in host cells. All viruses use the intracellular-trafficking machinery of the host to enter, replicate in and exit host cells. After uptake, bacteria such as Shigella spp. and Listeria spp. immediately lyse the vacuoles in which they are present, resulting in the release of these bacteria into the cytosol, where they replicate and spread to neighbouring cells. By contrast, other intracellular pathogens — for example, Mycobacterium spp., Salmonella spp., Chlamydia spp., Brucella spp., Legionella spp., Leishmania spp. and Toxoplasma spp. — are internalized into a membrane-bound vacuole and remain there. However, even among these pathogens, there are different strategies for generating an environment that is suitable for replication, and the pathogen-containing vacuoles show distinct characteristics: for example, Mycobacterium spp. prevent acidification of the phagosome and therefore do not interact with late endosomes or lysosomes, whereas Salmonella-containing vacuoles are characterized by the presence of several markers of late endocytic compartments. By contrast, other pathogens escape the endocytic pathway and interact with organelles of the secretory pathway: for example, Chlamydia spp. have been shown to intercept exocytic vesicles and acquire lipids from the trans-Golgi network. In this Review, we focus on pathogens that interact with the endoplasmic reticulum (ER) (in particular, viruses, Toxoplasma gondii, Brucella spp. and Legionella pneumophila), and we discuss the cellular and immunological consequences of this interaction. Interestingly, although the ER is an ideal location for pathogens to hide from the host immune system, emerging data indicate that several ER functions are important for the host to combat intracellular microorganisms.
Biology of the ER
The ER is a crucial intracellular organelle that is involved in the biosynthesis of cellular molecules, including proteins, carbohydrates and lipids. These become constituents of the ER or are transported to the Golgi, plasma membrane, endosomes and lysosomes. Regarding the immune system, the ER is the main site for the assembly of molecular complexes that are involved in antigen presentation by MHC class I and class II molecules: for example, it is the site of peptide loading onto MHC class I molecules.
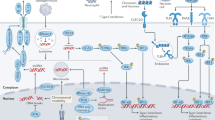
ER–Golgi biogenesis. The ER controls the routing of newly synthesized proteins that are destined for the exocytic pathway (that is, transport to the Golgi, plasma membrane or extracellular fluid) and the endocytic pathway (that is, transport to the endosomes or lysosomes), as well as those that are destined for the phagosomes and autophagosomes. The regulatory machinery that is present in the ER operates at multiple steps to control the routing of a particular protein. The first step towards exit from the ER is the formation of functional ER-exit sites, from which vesicles that contain the cargo protein bud off. These vesicles then form the ER–Golgi intermediate compartment (ERGIC), which in turn gives rise to the Golgi (Fig. 1).
a | Activation of SAR1 leads to the recruitment of the components of the coat complex COPII to endoplasmic reticulum (ER) membranes. COPII recruits cargo proteins that are to be delivered to other organelles and packages these into membrane tubules and vesicles. Activation of ADP-ribosylation factor 1 (ARF1) leads to recruitment of the components of the coat complex COPI to early secretory vesicles. On these membranes, exchange of the COPII coat for a COPI coat allows further protein sorting to occur and allows transport of cargo proteins away from the ER. Cells can then assemble a functional Golgi apparatus, and the secretory pathway is fully operational. b | In cells that express a particular mutant SAR1 protein (in which a histidine residue is replaced by a glycine residue at position 79) or a particular mutant ARF1 protein (in which a threonine residue is replaced by an asparagine residue at position 31), ER-exit sites are formed, but cargo is not transported away from the ER. c | In cells that express a different mutant SAR1 protein (in which a threonine residue is replaced by an asparagine residue at position 39), recruitment of the components of COPII to membranes is inhibited, and ER-exit sites do not form. ERGIC, endoplasmic-reticulum–Golgi intermediate compartment.
The formation of ER-exit sites is a complex process that involves recruitment of the components of the coat complex COPII to the ER membrane and polymerization of the coat complex on the buds, and both recruitment and polymerization are regulated by the small GTPase SAR1 (Refs 1, 2) (Fig. 1). Other regulatory proteins that participate in assembly of COPII include SEC16 and SEC12, a guanine-nucleotide-exchange factor (GEF) for SAR1 (Ref. 3) (Box 1). Diversity in the subunits of COPII endows the coat with the ability to sort different cargo proteins and to regulate the sorting of these proteins differentially4,5. However, COPII-coated regions of the ER are not the only regions where protein export to the secretory pathway occurs. Indeed, protein cargo is also exported by a process of tubule outgrowth from non-coated regions of ER-exit sites6. The vesicle-docking protein p115, which is present at ER-exit sites (both in COPII-coated regions and in tubule outgrowths) interacts with various proteins with known roles in ER-to-Golgi transport — including Golgi matrix protein 130 (GM130), GIANTIN, SNARE (soluble-N-ethylmaleimide-sensitive-factor accessory-protein receptor) proteins such as ERS24 (ER SNARE of 24 kDa; also known as SEC22B), the GTPase RAB1, and GBF1 (Golgi-specific brefeldin-A-resistance factor 1), which is the GEF for the small GTPase ADP-ribosylation factor 1 (ARF1) — to induce the differentiation of the nascent ER-exit sites into the ERGIC7. ARF1 coordinates the formation of the ERGIC (Box 1) by recruiting a large number of proteins, such as ankyrin, spectrin, tubulin and tubulin-associated proteins, in addition to the proteins that comprise the coat complex COPI. Together, these molecular complexes allow the ER-exit sites to be transformed first into the ERGIC and then into Golgi membranes.
Importantly, all of these cellular mechanisms are involved in transporting MHC complexes from the ER to the cell surface, as well as in regulating the secretion of soluble immune mediators, such as chemokines and other cytokines. As a result, any perturbation in ER–Golgi biogenesis affects antigen presentation and immune effector functions. So, it is not surprising that the ER is a target for pathogens and that the interaction of pathogens with the ER results in modulation of subsequent immune responses.
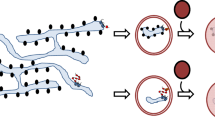
ER–phagocytosis interplay. Phagocytosis of inert particles, apoptotic bodies and invading pathogens is an essential part of generating an effective immune response. The ingested particles are degraded and processed to generate antigenic fragments for presentation by MHC molecules. During phagocytosis, cell signalling that leads to remodelling of the cytoskeleton results in the internalization of a particle into a membrane-bound organelle known as the phagosome. How phagosomes are formed is under debate. One hypothesis is that phagosomes are formed by invagination and fission of the plasma membrane (Fig. 2). A second hypothesis is that the ER has a key role in the formation of phagosomes; this process has been called ER phagocytosis (Fig. 2c). Support for this model is provided by the finding that, in macrophages, the ER has been shown to be recruited to the site of internalization of latex beads, Salmonella enterica serovar Typhimurium (S. typhimurium) and Leishmania donovani8. Furthermore, mutant Dictyostelium discoideum that lack both calreticulin and calnexin, the two main ER-resident proteins, have reduced phagocytic activity9. More-recent findings, by Becker et al.10, have also shown that impaired function of ERS24, a SNARE protein that mediates fusion of ER-derived vesicles with their target membrane and that is involved in the transition between the ER and the ERGIC, resulted in reduced engulfment of 3 μm particles by the J774 macrophage cell line. However, the ER-phagocytosis model is still controversial, and a recent study failed to detect continuity between the plasma membrane and ER components during the onset of phagocytosis11. In addition, despite the observation that the ER seems to be involved in phagosome biogenesis, ER-mediated phagocytosis is not used by Brucella abortus or Mycobacterium avium to enter host cells11. In conclusion, ER–phagosome interactions are complex and might depend on the intrinsic properties of the ingested particles. In the case of pathogens, expression of virulence factors that are involved in ER recruitment might be important in determining the intracellular fate of the pathogen.
a | In the first model, which is the classical model of phagocytosis and phagosome maturation, the plasma membrane invaginates and fuses, engulfing the extracellular particles and generating a phagosome. The phagosome then undergoes sequential interactions with both early and late endosomes, during which it acquires MHC class II molecules that are ready for peptide loading. b | In a second model of phagocytosis and phagosome maturation, in addition to interacting with the early and late endosomes, the plasma-membrane-derived phagosome undergoes several waves of fusion and fission with endoplasmic reticulum (ER)-derived vesicles. This could account for the delivery of MHC class I molecules and associated molecules to the phagosome and would facilitate cross-presentation of exogenously derived antigens. c | In a third model, recruitment of the ER to the site of phagocytosis provides MHC class I molecules and associated molecules at the beginning of the phagocytic process. The MHC class I molecules are retained in the phagosome until they are loaded with specific antigens that have been processed in the phagosome.
Newly formed phagosomes undergo a process of maturation that includes membrane fusion and fission with other intracellular organelles, such as endosomes and Golgi-derived vesicles. In addition, as they mature, phagosomes seem to acquire ER markers transiently through the fusion and fission of several waves of ER membranes8 (Fig. 2). Cross-presentation by dendritic cells (DCs) has been under discussion for the past two decades. However, recent work provides the basis of a framework for understanding how antigens that are derived from internalized material can be presented by MHC class I molecules12,13,14. Until now, cross-presentation of most peptide antigens has been thought to involve the transfer of antigens from the lumen of endocytic and phagocytic compartments to the cytosol, although the mechanism of this transfer has not been determined. After they reach the cytosol, antigens are digested by proteasomal proteolysis, and the resultant peptides are translocated to the ER by the ATP-dependent heterodimer TAP (transporter associated with antigen processing)15. These peptides are then loaded onto MHC class I molecules at the inner ER membrane. Because ER membranes are present in phagosomes, cross-presentation might occur directly in the phagosome without the need for antigen transfer. Consistent with this, proteasomes have also been found in association with latex-bead-containing phagosomes, ensuring that all of the cross-presentation machinery is in close association with the phagosome12. However, even though this model is attractive, retrograde vesicle-mediated transport from phagosomes to the Golgi and then to the ER cannot be ruled out16. Finally, the outcome of the maturation pathway is fusion of phagosomes with lysosomes. This allows acidification of the phagosome lumen, acquisition of lysosomal enzymes and generation of reactive oxygen intermediates (such as superoxide, which is produced through the reduction of molecular oxygen by NADPH oxidase)17.
Microbial pathogens interact with the ER
Intracellular pathogens have evolved to interact with specific intracellular organelles, allowing these pathogens to establish their replication niche. The ER is one of the organelles of the host cell that is targeted by intracellular pathogens. Several types of virus, parasite and bacterium have distinct mechanisms that use this organelle to support intracellular growth and, in many cases, to interfere with host immunity.
Viruses. Viruses are obligate intracellular organisms that require the machinery of the host cell to translate viral RNA into proteins, to replicate the viral genome and to produce large numbers of infectious viral particles. Numerous studies have been carried out to identify and understand how some viral proteins target the ER. Here, we focus on examples of human viruses that use ER membranes for translation, replication, and/or budding of viral particles, often resulting in the alteration of cellular membrane transport and therefore host immunity.
Hepatitis C virus (HCV), a member of the Flaviviridae family, is a single-stranded RNA virus. Infection with HCV causes an acute disease that can develop into a chronic disease and lead to severe complications, such as hepatocellular carcinoma18. The genome of HCV encodes a single polyprotein, which is proteolytically cleaved and processed by cellular and viral enzymes into several structural and non-structural proteins, and these proteins then contribute to viral replication19. Replication of RNA viruses occurs in a membrane-bound viral-replication complex that is composed of RNA and both viral and cellular proteins. In the case of HCV, the replication cycle is still not well understood. It seems that, after entry to the host cell, the RNA genome is released into the cytosol and translated, and the viral proteins that are generated after processing of the single polyprotein remain tightly associated with cellular membranes to form replicase complexes. New RNA molecules can then be produced, enabling further polyprotein expression and virion packaging. Several studies have shown that RNA replication occurs in perinuclear ER-bound membrane complexes20, although it also been suggested that HCV replication complexes are associated with detergent-resistant membranes in the Golgi21. Evidence for the former comes from the observation that several viral proteins that are involved in the replication of HCV RNA target the ER when expressed using heterologous systems19,20. In addition, expression of the complete HCV polyprotein or the HCV protein NS4B (non-structural protein 4B) alone resulted in changes to the morphology of the ER membrane, with the formation of a membranous web containing all of the viral structural and non-structural proteins22. These membranous structures, which are also present in cells that contain replicating HCV RNAs and in liver biopsies from HCV-infected chimpanzees, seem to be the site of synthesis of HCV RNA20. The production and processing of large amounts of viral proteins in ER-derived membranes disrupts normal ER function and induces considerable ER stress, which promotes the persistence of HCV in host cells23,24. HCV-induced ER stress also results in the disruption of protein folding and the accumulation of unfolded MHC class I molecules in the ER, with a subsequent reduction in cell-surface MHC class I expression24. Infection with HCV has also been shown to alter calcium homeostasis in the ER and to induce oxidative stress, both of which, together with ER stress, activate nuclear factor-κB and signal transducer and activator of transcription 3 (STAT3)25, thereby inducing the expression of genes that are involved in cellular processes that control inflammation, apoptosis and proliferation.
Several HCV proteins have themselves been shown to target signalling pathways that control important immune functions. For example, E2 (envelope protein 2) and NS5A inhibit the ER kinase PKR (interferon (IFN)-inducible double-stranded RNA-dependent protein kinase) without decreasing overall protein synthesis, and this represses the antiviral effect of IFNβ24,26,27. By contrast, the precursor protein NS4A–NS4B seems to affect the rate of ER-to-Golgi transport and to reduce cell-surface expression of MHC class I molecules28.
Other viruses specifically target components of the secretory pathway for use in the formation of infectious virions, and this results in disruption of ER-to-Golgi vesicle transport. One example is poliovirus, a member of the Picornaviridae family. Simultaneous with translation of poliovirus RNA into proteins, vesicles that contain newly synthesized RNA and viral proteins emerge from the ER and form nascent replication complexes. These vesicles seem to be formed in the ER using the machinery that forms COPII, as indicated by the presence of both SEC13 and SEC31 at the vesicular surface29. It is not clear whether formation of replication complexes derived from the ER has a direct effect on the immune response, but infection with poliovirus does result in inhibition of secretion of host proteins and in a reduction in antigen presentation30. This has been attributed to poliovirus protein 3A, which blocks ER-to-Golgi transport, leading to reduced secretion of cytokines (such as interleukin-6 (IL-6), CXC-chemokine ligand 8 (CXCL8; also known as IL-8) and IFNβ), decreased expression of tumour-necrosis-factor receptor (TNFR) at the cell surface and inhibition of antigen presentation by MHC class I molecules31,32,33,34,35. Because protein 3A is not essential for viral replication, it might provide a mechanism by which poliovirus evades immune recognition. Interestingly, protein 3A from another Picornaviridae-family enterovirus, coxsackievirus, was also shown to block the secretory pathway efficiently by blocking ER-to-Golgi transport36, but protein 3A from yet another Picornaviridae-family member (foot-and-mouth disease virus) did not35, highlighting the variety of mechanisms used by different viruses to replicate in host cells and to evade the immune response.
The herpesvirus human cytomegalovirus (CMV) has developed various mechanisms to evade the host immune response. Human CMV expresses several proteins that independently block various steps of the MHC class I antigen-presentation pathway, preventing T cells from recognizing the virus37,38. For example, the viral ER integral-membrane protein US6 binds TAP and inhibits peptide translocation to the ER and therefore peptide loading onto MHC class I molecules, whereas the ER-resident viral proteins US2 and US11 reroute MHC class I molecules to the cytosol, where they are degraded by the proteasome. The interactions of the human CMV protein US3 are more complex, and US3 has multiple functions in the cell. For example, it has been shown to bind transiently to peptide-loaded MHC class I molecules and, as a result of the presence of an ER-retention signal, it prevents MHC class I molecules from leaving the ER39. In addition, US3 binds tapasin, and this prevents peptide translocation to the ER and therefore peptide loading onto MHC class I molecules40. It also binds HLA-DR, inhibiting MHC class II molecules from presenting extracellular antigens taken up by phagocytic cells41.
Adenoviruses can also downregulate antigen presentation by MHC class I molecules. The protein E19, which binds MHC molecules, contains a dilysine motif that is recognized by COPI as an ER-retrieval signal. This ensures that E19 (and any molecules that are associated with E19) is recycled into the ER, thereby blocking transport of MHC class I molecules. E19 has also been shown to bind TAP and interfere with peptide loading onto MHC class I molecules in the ER37. These examples (which are summarized in Table 1) highlight several of the strategies that are used by viruses to block specific membrane transport along the endocytic pathway and that have important consequences for evasion of the host immune response by these viruses.
T. gondii. T. gondii is an obligate intracellular parasite that infects most nucleated cells of warm-blooded animals. In humans, T. gondii infection is one of the most common parasitic infections, but it causes severe disease only in congenitally infected neonates and immunocompromised individuals. During invasion of host cells, T. gondii directs sequential secretion of proteins from three specific types of secretory organelle, which are known as micronemes, rhoptries and dense granules42. First, micronemal proteins that contain adhesin domains are secreted, and these mediate recognition of, and attachment to, the host cell. And second, proteins that are secreted from the rhoptries, known as rhoptry proteins (ROPs), are involved in the formation of phagosomes. After T. gondii has entered the host cell, it resides in a specialized compartment known as the parasitophorous vacuole, where it quickly modifies the vacuolar membrane to remove host-cell components, thereby preventing fusion of the vacuole with endocytic compartments43,44. The parasitophorous vacuole therefore remains at neutral pH, and it forms extensive associations with the mitochondrion and ER of the host cell45,46. It is still unclear, however, how the parasite establishes an interaction with these organelles, but this process seems to involve further secretion of proteins from the rhoptries, as well as secretion of proteins from the dense granules47. For example, the T. gondii protein ROP2, which is known to contain mitochondria- and ER-targeting domains, has been shown to be secreted into the cytosol of the host cell, where it becomes phosphorylated48,49. Inactivation of ROP2 was found to result in abnormal parasitophorous vacuoles that were devoid of mitochondrial markers and contained reduced amounts of cholesterol but still associated with the ER50. Recently, the T. gondii protein GRA3 (dense-granule protein 3) was shown to contain an ER-retrieval motif, which might mediate the association of parasitophorous vacuoles with the ER51. Therefore, further work is required to determine which parasite proteins mediate interaction with the ER and whether association of the parasitophorous vacuole with the ER has any marked effects on the immune functions of host cells.
Brucella spp. Brucella spp. are Gram-negative bacteria that are facultative intracellular pathogens. They cause a zoonosis that leads to sterility and abortion in various mammals, including livestock and wild animals. In humans, disease is mainly caused by B. abortus, Brucella melitensis and Brucella suis, and this occurs after contact with infected animals or contaminated animal products. Human brucellosis is a chronic and debilitating illness with severe complications such as arthritis and endocarditis. Several virulence factors that are important for persistence of Brucella spp. in animal models or in in vitro cell cultures have been identified52,53,54.
Under non-opsonizing conditions, entry of bacteria to macrophages occurs through a mechanism that depends on lipid rafts, and the bacteria can therefore avoid extensive interaction with endocytic compartments55,56,57,58. Soon after a Brucella bacterium has been internalized, the Brucella-containing vacuole (BCV) has been shown to interact transiently with early endosomes, but it does not acquire late endosomal and lysosomal markers, such as RAB7, the mannose-6-phosphate receptor and cathepsin D55,58,59. Recent work has shown that Brucella spp. synthesize β-1,2-glucans, which modify membrane composition at the level of lipid rafts and prevent fusion with lysosomes60. Instead of fusing with lysosomes, BCVs fuse with the ER membrane and establish an ER-derived compartment in which Brucella can replicate58,61,62,63. During intracellular replication in both epithelial cells and macrophages, the BCV contains ribosomes and numerous ER markers such as the translocator SEC61 and the lectin chaperones calnexin and calreticulin. Confirmation of BCV fusion with the ER membrane comes from the immunocytochemical detection of glucose-6-phosphatase activity at the BCV membrane and the detection of ER markers in this location by electron microscopy58,63.
A recent study has shown that B. abortus cannot establish an ER-derived replicative compartment in cells that express a SAR1 variant that remains locked in the inactive (GDP bound) form64. This indicates that SAR1 activity and the subsequent recruitment of COPII to ER-exit sites is essential for the biogenesis of the ER-derived BCV (Fig. 3). It is probable that other components of the SAR1–COPII machinery are involved in fusion of BCVs with the ER membrane, but this remains to be investigated. On the part of the bacterium, there is strong evidence that the VirB type IV secretion system of Brucella spp. is required for BCV–ER fusion, because Brucella spp. with a mutated virB operon cannot sustain interactions with the ER membrane and, consequently, fail to replicate intracellularly58,65. Because such mutant bacteria are also attenuated in virulence in the mouse model of infection, the ability of B. abortus and B. melitensis to survive and replicate within a vacuole that interacts with the ER is essential for them to cause disease. It is possible that this important virulence system encodes specific secreted molecules that enable BCVs to fuse with the ER but that these molecules have not yet been identified. It remains unknown whether the establishment of an ER-derived replicative niche has consequences for the ability of host cells to mount an appropriate immune response to this pathogen. However, we recently showed that B. abortus lipopolysaccharide specifically inhibits MHC class II, but not MHC class I, antigen presentation53. These results are compatible with the immunosuppressive stages that have been observed in both mouse and human brucellosis66,67.
a | Viruses have evolved proteins that interfere with TAP (transporter associated with antigen processing) heterodimers in the lumen of the endoplasmic reticulum (ER): for example, the adenovirus protein E19, the human cytomegalovirus (CMV) proteins US3 and US6, and the herpes simplex virus (HSV) protein ICP47. The adenovirus protein E19 also interferes with retrograde transport, from the Golgi to the ER, and causes peptide-loaded MHC class I molecules to be returned to the ER. By contrast, several other proteins — such as the human CMV proteins US3 and US10, the hepatitis C virus (HCV) precursor protein non-structural protein 4A (NS4A)–NS4B, poliovirus protein 3A and coxsackievirus protein 3A — interfere with transport from the ER to the Golgi. b | Brucella-containing vacuoles require the activity of SAR1 for productive interactions with the ER. Cells that express a mutant SAR1 protein in which a threonine residue is replaced by an asparagine residue at position 39 (SAR1T39N) do not allow the replication of Brucella abortus. By contrast, Legionella pneumophila-containing vacuoles require the sequential activity of SAR1 and ADP-ribosylation factor 1 (ARF1) for productive interactions with the ER. Cells that express either a mutant SAR1 protein in which a histidine residue is replaced by a glycine residue at position 79 (SAR1H79G) or a mutant ARF1 protein in which a threonine residue is replaced by an asparagine residue at position 31 (ARF1T31N) do not allow replication of L. pneumophila. In the case of Toxoplasma gondii, the T. gondii proteins dense-granule protein 3 (GRA3) and rhoptry protein 2 (ROP2) might be involved in mediating interactions between the parasitophorous vacuole of T. gondii and the ER of the host. ERGIC, ER–Golgi intermediate compartment; β2m, β2-microglobulin.
L. pneumophila. L. pneumophila is a Gram-negative bacterium that is commonly found in aquatic environments, where it replicates inside protozoan hosts68. Aerosols that contain L. pneumophila can lead to respiratory infections in humans, and these infections can result in a severe pneumonia that is known as legionnaires' disease69. L. pneumophila replicates in mononuclear phagocytes70, and the main host cells for propagation of L. pneumophila are likely to be alveolar macrophages in the lungs71. In early investigations of the interactions of L. pneumophila with host cells, it was observed by electron microscopy that the L. pneumophila-containing vacuole (LCV) is different from the vacuoles that contain most other intracellular bacteria72,73. The two main features that are unique to the LCV are the absence of fusion with lysosomes and the presence of ribosomes surrounding the vacuolar membrane, and these features indicate that L. pneumophila can alter transport of the vacuole.
Genetic analysis of L. pneumophila mutants that cannot replicate in macrophages uncovered a specialized secretion system, known as the Dot/Icm system, that is required for the evasion of phagosome–lysosome fusion and for the generation of the ribosome-covered organelles74,75,76,77. The Dot/Icm secretion system is used for the delivery of bacterial proteins to the cytosol of eukaryotic cells. When the morphology of vacuoles that contain wild-type L. pneumophila is compared with vacuoles that contain mutant L. pneumophila with a defective Dot/Icm system, several differences can be observed. Vesicles attach and flatten along the limiting membrane of vacuoles that contain wild-type L. pneumophila, whereas vacuoles that contain L. pneumophila mutants with a non-functional Dot/Icm system are devoid of these attached vesicles78. The icmS and icmW genes of L. pneumophila encode accessory proteins that facilitate efficient transport of bacterial substrates into host cells by the Dot/Icm system. Interestingly, L. pneumophila mutants in which icmS or icmW are mutated reside in vacuoles that fuse rapidly and efficiently with lysosomes; however, vesicles are similarly recruited and attach to the vacuoles that contain these mutant bacteria78,79,80, indicating that attachment of host vesicles to LCVs can occur independently of evasion of lysosome fusion by L. pneumophila.
Several lines of evidence indicate that the vesicles attached to LCVs are derived from the host ER and that the Dot/Icm system enables L. pneumophila to intercept these vesicles as they transit between the ER and the Golgi. ER markers, such as the chaperone protein immunoglobulin-heavy-chain-binding protein (BIP), have been localized to the attached vesicles81,82, and the membrane thickness of the attached vesicles is similar to that of the ER membrane78. ARF1, RAB1 and ERS24, which are host factors that regulate membrane transport and fusion between the ER and the Golgi, also localize to vacuoles that contain wild-type L. pneumophila, but these proteins are absent from vacuoles that contain L. pneumophila with mutated Dot or Icm genes83,84,85 (Fig. 3). This indicates that these three proteins are important for biogenesis of an organelle that supports replication of L. pneumophila. This has been more conclusively shown in studies in which inhibitors that interfere with the functioning of ARF1 and RAB1 not only disrupt the transport of host secretory vesicles but also adversely affect intracellular replication of L. pneumophila83,84. In addition, it has been shown that L. pneumophila proteins that are translocated to the cytosol of the host cell by the Dot/Icm system activate the small GTPases ARF1 and RAB1 by functioning as GEFs86, indicating that L. pneumophila has evolved a specific mechanism to modulate the function of these host factors.
From these studies, a model has emerged in which proteins are translocated to host cells by the Dot/Icm system, and these proteins confer on the LCV properties that are, in many ways, similar to properties of the Golgi. This allows L. pneumophila to recruit secretory vesicles travelling from the ER to the Golgi and to promote fusion of these vesicles with the LCV to remodel this plasma-membrane-derived organelle into a novel early secretory compartment. As a result, L. pneumophila is contained in a non-degradative organelle that is rich in peptides, which function as the main carbon source for growth of L. pneumophila.
It was thought that, by residing in a non-degradative organelle, L. pneumophila would be able to avoid degradation in lysosomes, thereby restricting the presentation of L. pneumophila-derived antigens by MHC class II molecules. Recent studies, however, have indicated that CD4+ T cells from mice that have been immunized with L. pneumophila respond better to antigen-presenting cells infected with virulent L. pneumophila than to antigen-presenting cells infected with avirulent L. pneumophila (with a mutated dotA locus) that is transported to lysosomes87. Interestingly, this increased CD4+ T-cell response requires the intracellular synthesis of L. pneumophila antigens, but it does not require replication of L. pneumophila. This finding indicates that a unique subset of L. pneumophila-derived peptides is presented by MHC class II molecules after bacteria have established an ER-derived organelle. It has been shown that processing of L. pneumophila antigens for presentation by MHC class II molecules occurs in acidified endocytic compartments88, indicating that antigens traffic out of the LCV in vesicles that fuse with lysosomes before presentation at the plasma membrane. It is also thought that antigens delivered to the host cytosol by the Dot/Icm system are processed for presentation by MHC class I molecules to CD8+ T cells; however, MHC-class-I-restricted antigens remain to be identified. Ultimately, T cells that recognize antigens presented at the cell surface of macrophages with L. pneumophila replicating in ER-derived vacuoles would be important for a protective cell-mediated immune response. Given the ability of L. pneumophila to manipulate the secretory pathway of the host cell, it will be interesting to determine whether bacteria that reside in macrophages can avoid immune detection by perturbing transport pathways that are used for secretion of cytokines and transport of MHC complexes to the cell surface.
ER functions involved in microbial defence
Although microbial pathogens have evolved mechanisms to subvert ER functions, thereby influencing the host immune response, new studies of signalling pathways and membrane transport that involves the ER are beginning to uncover novel functions for the ER in host protection against microbial pathogens and disease (Fig. 4).
Although microbial pathogens have evolved mechanisms to subvert endoplasmic reticulum (ER) functions, the ER also has an important role in combating microorganisms. For example, under starvation conditions or following treatment with interferon-γ (IFNγ), host cells that are infected with Mycobacterium tuberculosis trigger autophagy, and the ER contributes to the formation of the autophagosome. Autophagosomes can then fuse with lysosomes, resulting in the destruction of M. tuberculosis trapped inside autophagosomes. In addition, infection with a virus, such as herpes simplex virus, induces the ER stress response, and exposure to IFNs induces the upregulation of expression of antimicrobial factors in the ER, including viperin and the p47 GTPases IGTP (IFNγ-induced GTPase) and IIGP1 (IFN-inducible GTPase 1).
Autophagy. Autophagy is a normal metabolic process by which a cell can capture cytosolic material in membrane-bound vacuoles known as autophagosomes. Although the biogenesis of autophagosomes is still ill-defined, several lines of evidence show that autophagosomes are generated from assembly of pre-autophagosomal structures that require functional ER and Golgi elements, such as the transport-protein particle complex and components of COPII, which have an important role in vesicular transport89,90,91. Autophagosomes are 'self-eating' compartments that undergo a process of maturation that leads to degradation of their contents after fusion with lysosomes. Recent studies have shown that, under conditions ranging from starvation to hormone treatment, several pathogenic bacteria transiently reside in autophagosomes63,92,93,94,95,96,97, and this might be an important step in the formation of a vacuole that supports pathogen replication98. In addition, recent studies have shown that viruses, including poliovirus, rhinoviruses and coronaviruses, interact with the autophagy machinery to undergo replication99,100.
Although, under certain circumstances, some viruses benefit from the formation of autophagosomes, it seems that host cells use autophagy to combat bacterial pathogens. For example, Mycobacterium tuberculosis is a facultative intracellular pathogen that resides in immature phagosomes and blocks phagosome–lysosome fusion. However, under conditions of starvation or after treatment with IFNγ, host cells that are infected with M. tuberculosis trigger autophagy. Consequently, phagosomes that contain M. tuberculosis are targeted by the autophagy machinery, resulting in the presence of autophagic vacuoles that contain M. tuberculosis in these cells. These autophagic vacuoles fuse with lysosomes, resulting in the destruction of M. tuberculosis trapped inside autophagosomes101. Unsurprisingly, pathogens have also developed strategies to resist the autophagy machinery. For example, Shigella flexneri is a Gram-negative bacterium that uses a type III secretion system to export bacterial effectors to the cytosol of the host cell. Among the secreted effectors, IcsB has been shown to prevent the autophagy machinery from recognizing S. flexneri in the cytosol. In the absence of the IcsB protein, the autophagy protein ATG5 (autophagy-related gene 5) can recognize the S. flexneri protein VirG and initiate autophagy. Indeed, when IcsB is absent, S. flexneri becomes trapped in autophagosomes, and bacterial survival is affected; however, the same mutant S. flexneri can survive in host cells that are deficient in ATG5 (Ref. 97). This shows that, although host cells use autophagy to eliminate pathogens, some pathogens have evolved mechanisms to avoid autophagy.
ER stress response. The ER contains chaperone proteins and modifying enzymes that assist in the process of protein folding. The accumulation of unfolded proteins in the ER can induce a stress response that leads to upregulation of expression of factors that are important for eliminating incorrectly folded proteins from the ER and for downregulating protein synthesis102. Recent studies indicate that infection with several viruses can induce ER stress and trigger an 'unfolded-protein response'24,103,104. Although the upregulation of expression of chaperone proteins that assist in protein folding could be beneficial for the assembly of viral particles, other aspects of the ER stress response are unfavourable for maintaining infection. For example, for infection with herpes simplex virus (HSV), inhibition of protein synthesis could be detrimental to viral propagation. HSV infection results in accumulation of viral proteins in the ER, and this triggers an ER stress response that involves activation of PERK (PKR-like EIF2α kinase)105. When activated, PERK phosphorylates eukaryotic translation-initiation factor 2α (EIF2α), and this phosphorylation interferes with translation of mRNA, thereby downregulating viral-protein synthesis. However, the HSV protein γ134.5 has been shown to dephosphorylate EIF2α, effectively counteracting the activity of PERK and therefore enabling the synthesis of viral proteins to continue105. A screen for small molecules that can block cell death resulting from prolonged ER stress identified a compound that is known as salubrinal, which functions as a selective inhibitor of cellular complexes that dephosphorylate EIF2α106. Interestingly, salubrinal was found to block the dephosphorylation of EIF2α that is induced by γ134.5, and the treatment of cells with salubrinal inhibited replication of HSV, showing that the ER stress response induced by HSV infection is detrimental to viral replication under conditions in which viral products cannot interfere with the ER-stress-response pathway106.
Another manifestation of ER stress is the induction of apoptosis107. Mouse caspase-12 and human caspase-4 localize to the ER membrane and have been shown to have an important role in triggering apoptotic cell death in response to ER stress108,109. It is possible that activation of caspase-12 and caspase-4 could also occur under conditions in which microbial pathogens have subverted ER functions. Induction of apoptosis in response to microbial subversion of ER functions would enable cells to limit pathogen growth by prematurely terminating the replication of the pathogen and by promoting phagocytosis of pathogens present in the apoptotic cells.
IFN-inducible factors. IFNs are cytokines that are produced in response to microbial infection and that have an important role in host defence against most intracellular pathogens. IFNγ is produced mainly by T cells and natural killer cells, and type I IFNs (in particular, IFNα and IFNβ) are produced by many cell types. The expression of several important immune mediators is upregulated in response to IFNs. Among these are proteins that localize to the ER and have an important role in restricting the growth of intracellular pathogens.
The protein viperin (also known as CIG5) was initially identified as a molecule that is upregulated in cells that are productively infected with human CMV110,111,112. Expression of viperin is induced in cells that are infected with human CMV, owing to the production of type I IFNs112. In uninfected cells, type-I-IFN-induced viperin is localized to the ER membrane because of the membrane-localization domain that is present near the amino terminus of viperin. However, when cells are infected with human CMV, viperin relocalizes sequentially from the ER to the Golgi and then to cytoplasmic vesicles. Markers that are found on host endosomal and secretory vesicles do not colocalize with these cytoplasmic vesicles; however, the human CMV structural proteins glycoprotein B (gB) and pp28 colocalize with viperin in these cytoplasmic vesicles, leading to the suggestion that the relocalization of viperin to these sites is directed by the virus and that this movement might be a viral strategy to prevent viperin from antagonizing viral replication. Indeed, if viperin expression is induced in cells before viral infection, viperin remains associated with the ER and interferes with the replication of human CMV112. It has been suggested that viperin might prevent the translation of viral products or interfere with the trafficking of viral components out of the ER; however, this remains to be shown.
The p47 GTPases are a recently identified family of proteins. Similar to viperin, expression of these proteins is induced by IFNs, and they have an important role in host defence against intracellular pathogens113,114. In mice, six p47-GTPase-family members have been cloned and partially characterized115,116,117,118,119,120. These proteins associate with distinct intracellular compartments. Similar to RAB GTPases, it is thought that, when p47 GTPases are in an active (GTP bound) state, they control membrane-transport processes, by recruiting effectors that promote endocytic maturation and the fusion of lysosomes with vacuoles that contain microbial pathogens. Three of the p47-GTPase-family members have been shown to associate with cellular membranes in uninfected cells. Two of these, IGTP (IFNγ-induced GTPase) and IIGP1 (IFN-inducible GTPase 1), localize to the ER, and the third, LRG47 (lipopolysaccharide- and IFN-induced GTPase of 47 kDa), is mainly found to be associated with membranes of the Golgi121,122,123. IGTP and LRG47 have both been shown to have a role in protection against infection with the protozoan pathogen T. gondii124,125,126, which creates a vacuole that is closely associated with the ER (as discussed earlier). LRG47 has also been implicated in host immunity to M. tuberculosis127. LRG47 can promote the fusion of lysosomes with a stalled early endosomal vacuole that contains M. tuberculosis, resulting in bacterial killing127. Recent evidence indicates that the recruitment of LRG47 to the vacuole that contains M. tuberculosis correlates with the recruitment of LC3, an effector of autophagy that is an elongation factor in autophagosome formation101. An intriguing possibility to explain this connection is that LRG47 facilitates maturation of vacuoles that contain M. tuberculosis by promoting the formation of an ER-derived autophagic organelle. So, it is possible that the p47 family of GTPases might be involved in promoting fusion of lysosomes with vacuoles that contain microbial pathogens by targeting these organelles for recognition by the autophagy pathway.
Concluding remarks
The challenge for the future is to characterize, at the molecular level, the interactions between bacterial products and host-cell targets. ER membranes are important components in the process of antigen presentation by MHC class I molecules to CD8+ T cells. Therefore, increasing our knowledge of ER-mediated interactions with pathogens will allow the manipulation of the immune response away from innate immunity towards adaptive immunity, especially towards a recognition programme that is based on CD8+ T-cell targeting of microbial epitopes. This, in turn, would push immunity towards cell-mediated killing of microorganisms.
Although the analysis of the complexity of the cellular and molecular pathways that are used to fight an infection is greatly benefiting from in vitro analysis, issues such as the generation of protective immunity can only be addressed through in vivo approaches. Therefore, the future is to develop innovative animal models to image the immune response to pathogens, with a focus on the function of specific target cells such as macrophages and DCs. For example, although some studies have already been initiated (such as those involving Legionella spp. and Salmonella spp.), further work is required to break down the complex in vivo process — including the contribution of all cellular processes and organelles — by which phagocytic cells, in particular DCs, differentiate in response to a pathogenic infection and elicit productive adaptive immune responses. Studies of pathogen–ER interactions have led to the discovery of new molecular mechanisms that bridge the phagocytic and the exocytic pathways and have opened new avenues for manipulating immunity to restrict pathogen growth and for identifying bacterial products that might be used as targets for the design of antimicrobial agents or vaccines.
References
Bonifacino, J. S. & Glick, B. S. The mechanisms of vesicle budding and fusion. Cell 116, 153–166 (2004).
Altan-Bonnet, N., Sougrat, R. & Lippincott-Schwartz, J. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16, 364–372 (2004).
Barlowe, C. et al. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907 (1994).
Roberg, K. J., Crotwell, M., Espenshade, P., Gimeno, R. & Kaiser, C. A. LST1 is a SEC24 homologue used for selective export of the plasma membrane ATPase from the endoplasmic reticulum. J. Cell Biol. 145, 659–672 (1999).
Shimoni, Y. et al. Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J. Cell Biol. 151, 973–984 (2000).
Mironov, A. A. et al. ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev. Cell 5, 583–594 (2003).
Garcia-Mata, R., Szul, T., Alvarez, C. & Sztul, E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum–Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell 14, 2250–2261 (2003).
Gagnon, E. et al. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110, 119–131 (2002). This article indicates that the ER membrane seems to become involved with phagocytic events at a very early stage and fuses with the phagocytic cup, leaving ER proteins orientated to the inner face of the phagosome.
Muller-Taubenberger, A. et al. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 20, 6772–6782 (2001).
Becker, T., Volchuk, A. & Rothman, J. E. Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc. Natl Acad. Sci. USA 102, 4022–4026 (2005).
Touret, N. et al. Quantitative and dynamic assessment of the contribution of the endoplasmic reticulum to phagosome formation. Cell 123, 157–170 (2005). This article highlights that the plasma membrane is the main constituent of nascent phagosomes, which then fuse with endosomes and, eventually, with lysosomes to become degradative organelles.
Houde, M. et al. Phagosomes are competent organelles for antigen cross-presentation. Nature 425, 402–406 (2003).
Guermonprez, P. et al. ER–phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425, 397–402 (2003). The phagosomes of DCs are organelles in which MHC class I molecules can be loaded with peptides that have been added exogenously. This article confirms the capacity of these phagosomes to carry out cross-presentation.
Desjardins, M. ER-mediated phagocytosis: a new membrane for new functions. Nature Rev. Immunol. 3, 280–291 (2003).
Watts, C. & Amigorena, S. Phagocytosis and antigen presentation. Semin. Immunol. 13, 373–379 (2001).
Sandvig, K. & van Deurs, B. Membrane traffic exploited by protein toxins. Annu. Rev. Cell Dev. Biol. 18, 1–24 (2002).
Vieira, O. V., Botelho, R. J. & Grinstein, S. Phagosome maturation: aging gracefully. Biochem. J. 366, 689–704 (2002).
Gremion, C. & Cerny, A. Hepatitis C virus and the immune system: a concise review. Rev. Med. Virol. 15, 235–268 (2005).
El-Hage, N. & Luo, G. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84, 2761–2769 (2003).
Moradpour, D. et al. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60, 103–109 (2003).
Aizaki, H., Lee, K. J., Sung, V. M., Ishiko, H. & Lai, M. M. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324, 450–461 (2004).
Egger, D. et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 (2002).
Pavio, N., Romano, P. R., Graczyk, T. M., Feinstone, S. M. & Taylor, D. R. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2α kinase PERK. J. Virol. 77, 3578–3585 (2003).
Tardif, K. D., Waris, G. & Siddiqui, A. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 13, 159–163 (2005).
Waris, G., Tardif, K. D. & Siddiqui, A. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER–nucleus signal transduction pathway and activates NF-κB and STAT-3. Biochem. Pharmacol. 64, 1425–1430 (2002).
Gale, M. J. Jr. et al. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230, 217–227 (1997).
Taylor, D. R., Shi, S. T., Romano, P. R., Barber, G. N. & Lai, M. M. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285, 107–110 (1999).
Konan, K. V. et al. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 77, 7843–7855 (2003).
Rust, R. C. et al. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75, 9808–9818 (2001).
Doedens, J. R. & Kirkegaard, K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14, 894–907 (1995).
Doedens, J. R., Giddings, T. H. Jr. & Kirkegaard, K. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71, 9054–9064 (1997).
Deitz, S. B., Dodd, D. A., Cooper, S., Parham, P. & Kirkegaard, K. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl Acad. Sci. USA 97, 13790–13795 (2000).
Neznanov, N. et al. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 75, 10409–10420 (2001).
Dodd, D. A., Giddings, T. H. Jr. & Kirkegaard, K. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and β interferon secretion during viral infection. J. Virol. 75, 8158–8165 (2001).
Choe, S. S., Dodd, D. A. & Kirkegaard, K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337, 18–29 (2005).
Wessels, E., Duijsings, D., Notebaart, R. A., Melchers, W. J. & van Kuppeveld, F. J. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 79, 5163–5173 (2005).
Hewitt, E. W. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110, 163–169 (2003).
Lybarger, L., Wang, X., Harris, M. & Hansen, T. H. Viral immune evasion molecules attack the ER peptide-loading complex and exploit ER-associated degradation pathways. Curr. Opin. Immunol. 17, 71–78 (2005).
Ahn, K. et al. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl Acad. Sci. USA 93, 10990–10995 (1996).
Park, B. et al. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 20, 71–85 (2004). This paper shows that the human CMV protein US3 interferes with the tapasin-mediated optimization of peptide binding to MHC class I molecules in the ER.
Hegde, N. R. et al. Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: a novel mechanism for evading major histocompatibility complex class II antigen presentation. J. Virol. 76, 10929–10941 (2002).
Binder, E. M. & Kim, K. Location, location, location: trafficking and function of secreted proteases of Toxoplasma and Plasmodium. Traffic 5, 914–924 (2004).
Mordue, D. G. & Sibley, L. D. Intracellular fate of vacuoles containing Toxoplasma gondii is determined at the time of formation and depends on the mechanism of entry. J. Immunol. 159, 4452–4459 (1997).
Mordue, D. G., Hakansson, S., Niesman, I. & Sibley, L. D. Toxoplasma gondii resides in a vacuole that avoids fusion with host cell endocytic and exocytic vesicular trafficking pathways. Exp. Parasitol. 92, 87–99 (1999).
Jones, T. C., Yeh, S. & Hirsch, J. G. The interaction between Toxoplasma gondii and mammalian cells. I. Mechanism of entry and intracellular fate of the parasite. J. Exp. Med. 136, 1157–1172 (1972).
Sinai, A. P., Webster, P. & Joiner, K. A. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J. Cell Sci. 110, 2117–2128 (1997).
Hakansson, S., Charron, A. J. & Sibley, L. D. Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 20, 3132–3144 (2001).
Sinai, A. P. & Joiner, K. A. The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J. Cell Biol. 154, 95–108 (2001).
Carey, K. L., Jongco, A. M., Kim, K. & Ward, G. E. The Toxoplasma gondii rhoptry protein ROP4 is secreted into the parasitophorous vacuole and becomes phosphorylated in infected cells. Eukaryot. Cell 3, 1320–1330 (2004).
Nakaar, V. et al. Pleiotropic effect due to targeted depletion of secretory rhoptry protein ROP2 in Toxoplasma gondii. J. Cell Sci. 116, 2311–2320 (2003). This study shows that targeted depletion of ROP2 gene expression by RNAi results in the loss of association of the mitochondrion with the membrane of the parasitophorous vacuole and in attenuated virulence of T. gondii in mice.
Henriquez, F. L. et al. Toxoplasma gondii dense granule protein 3 (GRA3) is a type I transmembrane protein that possesses a cytoplasmic dilysine (KKXX) endoplasmic reticulum (ER) retrieval motif. Parasitology 131, 169–179 (2005).
Roop, R. M., Bellaire, B. H., Valderas, M. W. & Cardelli, J. A. Adaptation of the brucellae to their intracellular niche. Mol. Microbiol. 52, 621–630 (2004).
Lapaque, N., Moriyon, I., Moreno, E. & Gorvel, J. P. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8, 60–66 (2005).
Fretin, D. et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell. Microbiol. 7, 687–698 (2005).
Pizarro-Cerda, J., Moreno, E., Sanguedolce, V., Mege, J. L. & Gorvel, J. P. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect. Immun. 66, 2387–2392 (1998).
Arenas, G. N., Staskevich, A. S., Aballay, A. & Mayorga, L. S. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 68, 4255–4263 (2000).
Naroeni, A., Jouy, N., Ouahrani-Bettache, S., Liautard, J. P. & Porte, F. Brucella suis-impaired specific recognition of phagosomes by lysosomes due to phagosomal membrane modifications. Infect. Immun. 69, 486–493 (2001).
Celli, J. et al. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198, 545–556 (2003).
Chaves-Olarte, E. et al. Activation of Rho and Rab GTPases dissociates Brucella abortus internalization from intracellular trafficking. Cell. Microbiol. 4, 663–676 (2002).
Arellano-Reynoso, B. et al. Cyclic β-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nature Immunol. 6, 618–625 (2005). This article shows that the B. abortus molecule β -1,2-glucan acts on the lipid rafts of vacuoles to prevent lysosomal fusion. This allows these pathogenic bacteria to reach the ER and survive.
Anderson, T. D. & Cheville, N. F. Ultrastructural morphometric analysis of Brucella abortus-infected trophoblasts in experimental placentitis. Bacterial replication occurs in rough endoplasmic reticulum. Am. J. Pathol. 124, 226–237 (1986).
Detilleux, P. G., Deyoe, B. L. & Cheville, N. F. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58, 2320–2328 (1990).
Pizarro-Cerda, J. et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66, 5711–5724 (1998).
Celli, J., Salcedo, S. P. & Gorvel, J. P. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc. Natl Acad. Sci. USA 102, 1673–1678 (2005). This study shows that SAR1 has an important role in allowing B. abortus to create a specialized vacuole that allows intracellular replication.
Comerci, D. J., Martinez-Lorenzo, M. J., Sieira, R., Gorvel, J. P. & Ugalde, R. A. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3, 159–168 (2001).
Giambartolomei, G. H. et al. Diminished production of T helper 1 cytokines correlates with T cell unresponsiveness to Brucella cytoplasmic proteins in chronic human brucellosis. J. Infect. Dis. 186, 252–259 (2002).
Zhan, Y., Yang, J. & Cheers, C. Cytokine response of T-cell subsets from Brucella abortus-infected mice to soluble Brucella proteins. Infect. Immun. 61, 2841–2847 (1993).
Fields, B. S. The molecular ecology of legionellae. Trends Microbiol. 4, 286–290 (1996).
McDade, J. E. et al. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory diseases. N. Engl. J. Med. 297, 1197–1203 (1977).
Horwitz, M. A. & Silverstein, S. C. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66, 441–450 (1980).
Nash, T. W., Libby, D. M. & Horwitz, M. A. Interaction between the legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J. Clin. Invest. 74, 771–782 (1984).
Horwitz, M. A. Formation of a novel phagosome by the legionnaires' disease bacterium (L egionella pneumophila) in human monocytes. J. Exp. Med. 158, 1319–1331 (1983).
Horwitz, M. A. The legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome lysosome fusion in human monocytes. J. Exp. Med. 158, 2108–2126 (1983).
Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 (1998).
Segal, G., Purcell, M. & Shuman, H. A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl Acad. Sci. USA 95, 1669–1674 (1998).
Marra, A., Blander, S. J., Horwitz, M. A. & Shuman, H. A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl Acad. Sci. USA 89, 9607–9611 (1992).
Berger, K. H. & Isberg, R. R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7, 7–19 (1993).
Tilney, L. G., Harb, O. S., Connelly, P. S., Robinson, C. G. & Roy, C. R. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114, 4637–4650 (2001).
Zuckman, D. M., Hung, J. B. & Roy, C. R. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol. Microbiol. 32, 990–1001 (1999).
Coers, J. et al. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38, 719–736 (2000).
Swanson, M. S. & Isberg, R. R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63, 3609–3620 (1995).
Abu Kwaik, Y. The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62, 2022–2028 (1996).
Kagan, J. C., Stein, M. P., Pypaert, M. & Roy, C. R. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199, 1201–1211 (2004). This study describes that L. pneumophila recruits the host GTPase RAB1 and the ER-derived vesicles that contain ERS24 (also known as SEC22B) to drive fusion of the bacterium-containing vacuole with the ER.
Kagan, J. C. & Roy, C. R. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nature Cell Biol. 4, 945–954 (2002).
Derre, I. & Isberg, R. R. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72, 3048–3053 (2004).
Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682 (2002).
Neild, A. L. & Roy, C. R. Immunity to vacuolar pathogens: what can we learn from Legionella? Cell. Microbiol. 6, 1011–1018 (2004).
Neild, A., Murata, T. & Roy, C. R. Processing and major histocompatibility complex class II presentation of Legionella pneumophila antigens by infected macrophages. Infect. Immun. 73, 2336–2343 (2005).
Reggiori, F. et al. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 2189–2204 (2004).
Ishihara, N. et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12, 3690–3702 (2001).
Meiling-Wesse, K. et al. Trs85 (Gsg1), a component of the TRAPP complexes is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280, 33669–33678 (2005).
Walker, D. H., Popov, V. L., Crocquet-Valdes, P. A., Welsh, C. J. & Feng, H. M. Cytokine-induced, nitric oxide-dependent, intracellular antirickettsial activity of mouse endothelial cells. Lab. Invest. 76, 129–138 (1997).
Sturgill-Koszycki, S. & Swanson, M. S. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192, 1261–1272 (2000).
Dorn, B. R., Dunn, W. A. Jr. & Progulske-Fox, A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 69, 5698–5708 (2001).
Rich, K. A., Burkett, C. & Webster, P. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5, 455–468 (2003).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004).
Ogawa, M. et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731 (2005). In this study, the type-III-secretion-system effector IcsB is shown to protect intracellular S. flexneri from autophagy by associating with the autophagosome protein ATG5 and inhibiting its binding to VirG.
Dunn, W. A. Jr. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 4, 139–143 (1994).
Jackson, W. T. et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3, e156 (2005).
Prentice, E., Jerome, W. G., Yoshimori, T., Mizushima, N. & Denison, M. R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279, 10136–10141 (2004).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Rutkowski, D. T. & Kaufman, R. J. A trip to the ER: coping with stress. Trends Cell Biol. 14, 20–28 (2004).
Li, X. D., Lankinen, H., Putkuri, N., Vapalahti, O. & Vaheri, A. Tula hantavirus triggers pro-apoptotic signals of ER stress in Vero E6 cells. Virology 333, 180–199 (2005).
Dimcheff, D. E., Faasse, M. A., McAtee, F. J. & Portis, J. L. Endoplasmic reticulum (ER) stress induced by a neurovirulent mouse retrovirus is associated with prolonged BiP binding and retention of a viral protein in the ER. J. Biol. Chem. 279, 33782–33790 (2004).
Cheng, G., Feng, Z. & He, B. Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2α dephosphorylation by the γ134.5 protein. J. Virol. 79, 1379–1388 (2005).
Boyce, M. et al. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307, 935–939 (2005). This study shows that treatment with salubrinal (an inhibitor of protein-phosphatase-1-mediated dephosphorylation of the translation-initiation factor EIF2α) protects cells from ER stress that is caused by the accumulation of unfolded proteins.
Szegezdi, E., Fitzgerald, U. & Samali, A. Caspase-12 and ER-stress-mediated apoptosis: the story so far. Ann. NY Acad. Sci. 1010, 186–194 (2003).
Nakagawa, T. et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403, 98–103 (2000).
Hitomi, J. et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. J. Cell Biol. 165, 347–356 (2004).
Boyle, K. A., Pietropaolo, R. L. & Compton, T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19, 3607–3613 (1999).
Zhu, H., Cong, J. P. & Shenk, T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl Acad. Sci. USA 94, 13985–13990 (1997).
Chin, K. C. & Cresswell, P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl Acad. Sci. USA 98, 15125–15130 (2001).
Taylor, G. A., Feng, C. G. & Sher, A. p47 GTPases: regulators of immunity to intracellular pathogens. Nature Rev. Immunol. 4, 100–109 (2004).
MacMicking, J. D. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25, 601–609 (2004).
Gilly, M. & Wall, R. The IRG-47 gene is IFN-γ induced in B cells and encodes a protein with GTP-binding motifs. J. Immunol. 148, 3275–3281 (1992).
Carlow, D. A., Marth, J., Clark-Lewis, I. & Teh, H. S. Isolation of a gene encoding a developmentally regulated T cell-specific protein with a guanine nucleotide triphosphate-binding motif. J. Immunol. 154, 1724–1734 (1995).
Lafuse, W. P., Brown, D., Castle, L. & Zwilling, B. S. Cloning and characterization of a novel cDNA that is IFN-γ-induced in mouse peritoneal macrophages and encodes a putative GTP-binding protein. J. Leukoc. Biol. 57, 477–483 (1995).
Sorace, J. M., Johnson, R. J., Howard, D. L. & Drysdale, B. E. Identification of an endotoxin and IFN-inducible cDNA: possible identification of a novel protein family. J. Leukoc. Biol. 58, 477–484 (1995).
Taylor, G. A. et al. Identification of a novel GTPase, the inducibly expressed GTPase, that accumulates in response to interferon γ. J. Biol. Chem. 271, 20399–20405 (1996).
Carlow, D. A., Teh, S. J. & Teh, H. S. Specific antiviral activity demonstrated by TGTP, a member of a new family of interferon-induced GTPases. J. Immunol. 161, 2348–2355 (1998).
Martens, S. et al. Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: retargeting to plasma membrane induced by phagocytosis. J. Immunol. 173, 2594–2606 (2004).
Zerrahn, J., Schaible, U. E., Brinkmann, V., Guhlich, U. & Kaufmann, S. H. The IFN-inducible Golgi- and endoplasmic reticulum-associated 47-kDa GTPase IIGP is transiently expressed during listeriosis. J. Immunol. 168, 3428–3436 (2002).
Taylor, G. A. et al. The inducibly expressed GTPase localizes to the endoplasmic reticulum, independently of GTP binding. J. Biol. Chem. 272, 10639–10645 (1997).
Taylor, G. A. et al. Pathogen-specific loss of host resistance in mice lacking the IFN-γ-inducible gene IGTP. Proc. Natl Acad. Sci. USA 97, 751–755 (2000).
Collazo, C. M. et al. The function of γ interferon-inducible GTP-binding protein IGTP in host resistance to Toxoplasma gondii is Stat1 dependent and requires expression in both hematopoietic and nonhematopoietic cellular compartments. Infect. Immun. 70, 6933–6939 (2002).
Collazo, C. M. et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon γ-inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194, 181–188 (2001).
MacMicking, J. D., Taylor, G. A. & McKinney, J. D. Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science 302, 654–659 (2003). This study shows that the activity of the GTPase LRG47 is responsible for the acidification of M. tuberculosis -containing vacuoles following activation by IFNγ.
Fromme, J. C. & Schekman, R. COPII-coated vesicles: flexible enough for large cargo? Curr. Opin. Cell Biol. 17, 345–352 (2005).
Altan-Bonnet, N., Phair, R. D., Polishchuk, R. S., Weigert, R. & Lippincott-Schwartz, J. A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis. Proc. Natl Acad. Sci. USA 100, 13314–13319 (2003).
Acknowledgements
C.R.R. is supported by the National Institutes of Health (USA). J.-P.E.G. and S.P.S. are supported by grants from the lnstitut National de la Santé et de la Recherche Médicale and the Centre National de la Recherche Scientifique (INSERM-CNRS) (France).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Autophagosome
-
An intracytoplasmic vacuole that contains components of the cytoplasm. It fuses with a lysosome to form an autophagolysosome, thereby subjecting its contents to enzymatic digestion.
- Small GTPase
-
A small (20–25 kDa) protein that binds GTP and functions as a molecular switch in intracellular-signalling pathways. Activation and deactivation of small GTPases can be considered to occur in a cycle, between the GTP-bound form (which is active) and the GDP-bound form (which is inactive). This cycle is controlled by other regulatory proteins.
- Cross-presentation
-
The ability of certain antigen-presenting cells to load peptides that are derived from exogenous antigens onto MHC class I molecules. This property is atypical. Most cells present peptides that are derived exclusively from their endogenous proteins on MHC class I molecules. Cross-presentation is required for the initiation of immune responses to viruses that do not infect antigen-presenting cells.
- Detergent-resistant membranes
-
Membrane domains that are enriched in cholesterol, phospholipids and sphingolipids. These domains are liquid ordered and insoluble in non-ionic detergents.
- ER stress
-
(Endoplasmic-reticulum stress). A response by the ER that results in disruption of protein folding and in accumulation of unfolded proteins in the ER.
- Picornaviridae
-
The picornavirus family. The name picornavirus means small RNA virus. These viruses are non-enveloped, positive-stranded RNA viruses with an icosahedral capsid.
- Enterovirus
-
A virus that resides in the gastrointestinal tract.
- Adhesin domain
-
A domain that mediates adhesion to host determinants such as glycolipids, proteoglycans, cell-surface proteins and extracellular-matrix proteins.
- Lipid raft
-
A micro-aggregate of cholesterol and sphingomyelin that is thought to occur in the plasma membrane. These regions are also known as glycolipid-enriched membrane microdomains (GEMs) or detergent-insoluble glycolipid-enriched membrane microdomains (DIGs).
- Glucose-6-phosphatase activity
-
In this case, activity is determined by a cytochemical method that uses electron microscopy to detect glucose-6-phosphatase present in the endoplasmic reticulum.
- VirB type IV secretion system
-
The genome of Brucella spp. contains the virB operon. This operon has high homology with the virB operon of Agrobacterium spp. and encodes a well-characterized type IV secretion system.
- Dot/lcm system
-
A bacterial type IV secretion apparatus that is used by the intracellular pathogen Legionella pneumophila. This secretion apparatus is composed of proteins that are encoded by 24 Dot (defective organelle trafficking) and Icm (intracellular multiplication) genes. The Dot/Icm system translocates bacterial proteins to the cytosol of host cells, and these translocated proteins promote transport of endoplasmic-reticulum-derived vesicles to vacuoles that contain L. pneumophila. The dotA gene is essential for translocation of bacterial proteins to host cells by the Dot/Icm system.
- Type III secretion system
-
A syringe-like, multisubunit secretion apparatus that can transport bacterial proteins directly into a eukaryotic cell by injection.
- p47 GTPases
-
A family of proteins that is encoded by interferon-γ-regulated genes. These proteins have specific roles in host defence against pathogens.
- RAB GTPases
-
A group of small GTPases that belongs to the RAS superfamily. These proteins are generally involved in regulation of numerous signalling pathways that control cytoskeletal integrity, proliferation, cell adhesion, apoptosis and cell migration.
Rights and permissions
About this article
Cite this article
Roy, C., Salcedo, S. & Gorvel, JP. Pathogen–endoplasmic-reticulum interactions: in through the out door. Nat Rev Immunol 6, 136–147 (2006). https://doi.org/10.1038/nri1775
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri1775
This article is cited by
-
Organellar homeostasis and innate immune sensing
Nature Reviews Immunology (2022)
-
Super resolution microscopy and deep learning identify Zika virus reorganization of the endoplasmic reticulum
Scientific Reports (2020)
-
‘Candidatus Liberibacter asiaticus’ Accumulates inside Endoplasmic Reticulum Associated Vacuoles in the Gut Cells of Diaphorina citri
Scientific Reports (2017)
-
Using host-pathogen protein interactions to identify and characterize Francisella tularensis virulence factors
BMC Genomics (2015)
-
Emerging functions of the unfolded protein response in immunity
Nature Immunology (2014)