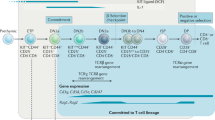
Key Points
-
The early, T-cell receptor (TCR)-independent stages of T-cell development are summarized in this Review. The process of specification, through which developing precursors gain T-cell characteristics, is paralleled by the process of commitment, through which they lose the potential to follow alternative developmental routes. Both of these processes now appear to occur in discrete steps, with an important transition within the double-negative 2 (DN2) stage.
-
Notch signalling is important from entry of T-cell precursors into the thymus to the culmination of T-cell-lineage commitment many cell divisions later. However, the gene regulatory effects of Notch signalling differ from stage to stage, greatly affected by shifts in the activities of other transcription factors and by responses to other environmental signals. Detailed expression patterns of multiple differentiation and regulatory genes are shown.
-
The initiation of the T-cell developmental programme by Notch signalling depends on the activity of at least four other transcription factors or transcription factor families which define the competence of pre-thymic precursors. Even when Notch signalling is under way, the differentiation of the early intrathymic precursors to the DN2 stage is controlled by a complex mixture of other microenvironmental signals and a newly defined negative feedback network for Notch itself.
-
A core group of transcription factors collaborates with Notch repeatedly throughout early T-cell development to maintain or advance T-cell-lineage differentiation. These are not turned on de novo during commitment, but rather are expressed steadily or at gently increasing levels from the earliest intrathymic stage to commitment. Other, crucial but less well studied, transcription factors are more dynamically upregulated at developmental transition points.
-
Another group of transcription factors, inherited from multipotent progenitors, is downregulated asynchronously and silenced during commitment. Factors in this non-T-cell-lineage class are apparently responsible for the lineage plasticity that early T-cell precursors retain for many cell generations within the thymus. One role of Notch signalling in vivo might be to constrain the lineage diversionary activities of this group of factors until they are permanently silenced.
-
The commitment process and the unleashing of T-cell-lineage differentiation gene expression are both correlated with downregulation of the non-T-cell-lineage factors. A critical component of the T-cell commitment mechanism that remains to be discovered is the repressor or repressor network that silences these non-T-cell-lineage factors.
Abstract
Multipotent blood progenitor cells enter the thymus and begin a protracted differentiation process in which they gradually acquire T-cell characteristics while shedding their legacy of developmental plasticity. Notch signalling and basic helix-loop-helix E-protein transcription factors collaborate repeatedly to trigger and sustain this process throughout the period leading up to T-cell lineage commitment. Nevertheless, the process is discontinuous with separately regulated steps that demand roles for additional collaborating factors. This Review discusses new evidence on the coordination of specification and commitment in the early T-cell pathway; effects of microenvironmental signals; the inheritance of stem-cell regulatory factors; and the ensemble of transcription factors that modulate the effects of Notch and E proteins, to distinguish individual stages and to polarize T-cell-lineage fate determination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hayday, A. C. & Pennington, D. J. Key factors in the organized chaos of early T cell development. Nature Immunol. 8, 137–144 (2007).
Anderson, M. K. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol. Rev. 209, 191–211 (2006).
Petrie, H. T. Cell migration and the control of post-natal T-cell lymphopoiesis in the thymus. Nature Rev. Immunol. 3, 859–866 (2003).
Rothenberg, E. V. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 10, 370–379 (2000).
Shortman, K. et al. The linkage between T-cell and dendritic cell development in the mouse thymus. Immunol. Rev. 165, 39–46 (1998).
Ceredig, R. & Rolink, T. A positive look at double-negative thymocytes. Nature Rev. Immunol. 2, 888–897 (2002). This review emphasizes the importance of using high KIT expression to define T-cell precursors in the DN1 population and to characterize DN2 cells. DN1 cells defined here largely correspond to the ETP DN1a and DN1b cells confirmed as precursors in references 25 and 131.
Blom, B. & Spits, H. Development of human lymphoid cells. Annu. Rev. Immunol. 24, 287–320 (2006).
Petrie, H. T. & Zuniga-Pflucker, J. C. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 25, 649–679 (2007).
Bhandoola, A., von Boehmer, H., Petrie, H. T. & Zuniga-Pflucker, J. C. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 26, 678–689 (2007).
Wu, L. T lineage progenitors: the earliest steps en route to T lymphocytes. Curr. Opin. Immunol. 18, 121–126 (2006).
Jenkinson, E. J., Jenkinson, W. E., Rossi, S. W. & Anderson, G. The thymus and T-cell commitment: the right niche for Notch? Nature Rev. Immunol. 6, 551–555 (2006).
Kawamoto, H. A close developmental relationship between the lymphoid and myeloid lineages. Trends Immunol. 27, 169–175 (2006).
Pelayo, R. et al. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr. Opin. Immunol. 17, 100–107 (2005).
Garbe, A. I. & von Boehmer, H. TCR and Notch synergize in αβ versus γδ lineage choice. Trends Immunol. 28, 124–131 (2007).
Boehm, T. & Bleul, C. C. Thymus-homing precursors and the thymic microenvironment. Trends Immunol. 27, 477–484 (2006).
Weerkamp, F., Pike-Overzet, K. & Staal, F. J. T. T-sing progenitors to commit. Trends Immunol. 27, 125–131 (2006).
Adolfsson, J. et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005).
Yoshida, T., Ng, S. Y., Zuniga-Pflucker, J. C. & Georgopoulos, K. Early hematopoietic lineage restrictions directed by Ikaros. Nature Immunol. 7, 382–391 (2006). This paper uses gene expression analysis and in vitro developmentto establish Ikaros functions in distinct types of multipotent haematopoietic progenitors.It shows that T-cell development qualitatively depends on Ikaros.
Garcia-Peydro, M., de Yebenes, V. G. & Toribio, M. L. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J. Immunol. 177, 3711–3720 (2006).
De Smedt, M., Hoebeke, I., Reynvoet, K., Leclercq, G. & Plum, J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood 106, 3498–3506 (2005).
Heinzel, K., Benz, C., Martins, V. C., Haidl, I. D. & Bleul, C. C. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J. Immunol. 178, 858–868 (2007).
Schwarz, B. A. et al. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 178, 2008–2017 (2007).
Masuda, K. et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 24, 4052–4060 (2005).
Yokota, T. et al. Unique properties of fetal lymphoid progenitors identified according to RAG1 gene expression. Immunity 19, 365–375 (2003).
Allman, D. et al. Thymopoiesis independent of common lymphoid progenitors. Nature Immunol. 4, 168–174 (2003).
Igarashi, H., Gregory, S. C., Yokota, T., Sakaguchi, N. & Kincade, P. W. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity 17, 117–130 (2002).
Taghon, T., Yui, M. A., Pant, R., Diamond, R. A. & Rothenberg, E. V. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006).
Schmitt, T. M. & Zuniga-Pflucker, J. C. Thymus-derived signals regulate early T-cell development. Crit. Rev. Immunol. 25, 141–160 (2005).
Maillard, I., Fang, T. & Pear, W. S. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23, 945–974 (2005).
Schmitt, T. M. & Zúñiga-Pflücker, J. C. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17, 749–756 (2002). By establishing a highly robust, accessible in vitro culture system for early T-cell development, this paper liberated T-cell development from the 'black box' of the thymic microenvironment and profoundly transformed the field.
Silva-Santos, B., Pennington, D. J. & Hayday, A. C. Lymphotoxin-mediated regulation of γδ cell differentiation by αβ T cell progenitors. Science 307, 925–928 (2005).
Gounari, F. et al. Tracing lymphopoiesis with the aid of a pTα-controlled reporter gene. Nature Immunol. 3, 489–496 (2002).
Masuda, K. et al. T cell lineage determination precedes the initiation of TCRβ rearrangement. J. Immunol. 179, 3699–3706 (2007). Using a pLCK–GFP transgene to mark a positive switchpoint in T-cell gene expression in living thymocytes, the authors discovered that the DN2 stage is composed of two distinct substages, and that DC developmental potential is lost by the time that the pLCK-GFP transgene is activated.
David-Fung, E. S. et al. Progression of regulatory gene expression states in fetal and adult pro-T cell development. Immunol. Rev. 209, 212–236 (2006).
Taghon, T. N., David, E.-S., Zúñiga-Pflücker, J. C. & Rothenberg, E. V. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 19, 965–978 (2005). This paper dissects the timing and gene expression signatures that correlate with lineage commitment in vitro in the presence or absence of Notch–DLL signals.
Balciunaite, G., Ceredig, R. & Rolink, A. G. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood 105, 1930–1936 (2005). This study shows that macrophage developmental potential is maintained by ETPs and DN2 thymocytes whereas B-cell potential is not. Macrophages generated from ETP and DN2 cells are phagocytically active and yet may contain limited TCRβ gene rearrangements, confirming their T-cell-lineage origin.
Tabrizifard, S. et al. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J. Immunol. 173, 1094–1102 (2004). This paper provides an expression analysis of transcription factor genes in murine pro-T cells, based on hybridization to Affymetrix microarrays. Hundreds of regulatory genes are tracked, and genes that are particularly useful for defining specific transitions are described.
Yui, M. A. & Rothenberg, E. V. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J. Immunol. 173, 5381–5391 (2004).
Chen, F., Rowen, L., Hood, L. & Rothenberg, E. V. Differential transcriptional regulation of individual TCR Vβ segments before gene rearrangement. J. Immunol. 166, 1771–1780 (2001).
Rothenberg, E. V., Diamond, R. A., Pepper, K. A. & Yang, J. A. Interleukin-2 gene inducibility in T cells prior to T-cell receptor expression: changes in signaling pathways and gene expression requirements during intrathymic maturation. J. Immunol. 144, 1614–1624 (1990).
Massa, S., Balciunaite, G., Ceredig, R. & Rolink, A. G. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur. J. Immunol. 36, 526–532 (2006).
Kang, J. & Der, S. D. Cytokine functions in the formative stages of a lymphocyte's life. Curr. Opin. Immunol. 16, 180–190 (2004).
King, A. G., Kondo, M., Scherer, D. C. & Weissman, I. L. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc. Natl Acad. Sci. USA 99, 4508–4513 (2002).
Benz, C. & Bleul, C. C. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J. Exp. Med. 202, 21–31 (2005).
Sambandam, A. et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nature Immunol. 6, 663–670 (2005).
Lu, M. et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J. Immunol. 175, 5848–5856 (2005).
Schmitt, T. M., Ciofani, M., Petrie, H. T. & Zúñiga-Pflücker, J. C. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J. Exp. Med. 200, 469–479 (2004).
Shen, H. Q. et al. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J. Immunol. 171, 3401–3406 (2003).
Wu, L., Li, C.-L. & Shortman, K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 184, 903–911 (1996). This paper first showed persistence of a non-lymphoid developmental option into the DN2 stage of thymocyte development, which is shut off in the DN3 stage.
Taghon, T., Yui, M. A. & Rothenberg, E. V. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nature Immunol. 8, 845–855 (2007). This paper shows that even the essential T-cell-lineage transcription factor GATA3 can act as a diversionary factor for ETP and DN2 T-cell precursors, if it is expressed at an elevated level. In this case Notch acts as an antagonist of GATA3 instead of a collaborator.
Rothenberg, E. V. Negotiation of the T lineage fate decision by transcription-factor interplay and micro-environmental signals. Immunity 26, 690–702 (2007).
Franco, C. B. et al. Notch/Delta signaling constrains re-engineering of pro-T cells by PU.1. Proc. Natl Acad. Sci. USA 103, 11993–11998 (2006). This paper details the regulatory cascade induced by PU.1 as it diverts thymocytes from the T-cell programme to the myeloid-cell or DC pathway. In DN2 and later thymocytes, Notch–DLL signalling precisely neutralizes a subset of PU.1 effects.
Tydell, C. C. et al. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J. Immunol. 179, 421–438 (2007). This study reports a panel of transcription factor and signalling genes that are upregulated in early T-cell-lineage development more than in precursors for other lineages. Most of the 'T-cell-lineage' genes are 'legacies' from stem or multipotent progenitors but Bcl11b is uniquely induced during the ETP to DN2 transition in vivo.
Tanigaki, K. & Honjo, T. Regulation of lymphocyte development by Notch signaling. Nature Immunol. 8, 451–456 (2007).
Reizis, B. & Leder, P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 16, 295–300 (2002).
Deftos, M. L., Huang, E., Ojala, E. W., Forbush, K. A. & Bevan, M. J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13, 73–84 (2000).
Maillard, I. et al. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 203, 2239–2245 (2006).
Rothenberg, E. V. Regulatory factors for initial T lymphocyte lineage specification. Curr. Opin. Hematol. 14, 322–329 (2007).
Rothenberg, E. V. & Taghon, T. Molecular genetics of T cell development. Annu. Rev. Immunol. 23, 601–649 (2005).
Staal, F. J. T., Weerkamp, F., Langerak, A. W., Hendriks, R. W. & Clevers, H. C. Transcriptional control of T lymphocyte differentiation. Stem Cells 19, 165–179 (2001).
Ho, I. C. & Pai, S. Y. GATA-3 – not just for Th2 cells anymore. Cell. Mol. Immunol. 4, 15–29 (2007).
Murre, C. Helix-loop-helix proteins and lymphocyte development. Nature Immunol. 6, 1079–1086 (2005).
Wakabayashi, Y. et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nature Immunol. 4, 533–539 (2003).
Wang, D. et al. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J. Immunol. 177, 109–119 (2006).
El Andaloussi, A. et al. Hedgehog signaling controls thymocyte progenitor homeostasis and differentiation in the thymus. Nature Immunol. 7, 418–426 (2006).
Goetz, T. L., Gu, T. L., Speck, N. A. & Graves, B. J. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol. Cell. Biol. 20, 81–90 (2000).
Gu, T. L., Goetz, T. L., Graves, B. J. & Speck, N. A. Auto-inhibition and partner proteins, core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1). Mol. Cell. Biol. 20, 91–103 (2000).
Melichar, H. J. et al. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science 315, 230–233 (2007).
Weerkamp, F. et al. Identification of Notch target genes in uncommitted T-cell progenitors: no direct induction of a T-cell specific gene program. Leukemia 20, 1967–1977 (2006).
Höflinger, S. et al. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 173, 3935–3944 (2004).
Schmitt, T. M. et al. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nature Immunol. 5, 410–417 (2004).
Huang, J. et al. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. J. Immunol. 175, 4858–4865 (2005).
Krueger, A., Garbe, A. I. & von Boehmer, H. Phenotypic plasticity of T cell progenitors upon exposure to Notch ligands. J. Exp. Med. 203, 1977–1984 (2006).
Lefort, N. et al. Short exposure to Notch ligand Delta-4 is sufficient to induce T-cell differentiation program and to increase the T cell potential of primary human CD34+ cells. Exp. Hematol. 34, 1720–1729 (2006).
Dakic, A. et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 201, 1487–1502 (2005).
Iwasaki, H. et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106, 1590–1600 (2005).
Talebian, L. et al. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood 109, 11–21 (2007).
Ikawa, T., Kawamoto, H., Goldrath, A. W. & Murre, C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 203, 1329–1342 (2006). The authors use an E2A -knockout haematopoietic progenitor cell line and tamoxifen to identify probable direct targets of E2A in a precursor-cell context. The results show synergistic and combinatorial interaction between Notch and E proteins that activate T-cell-lineage genes.
Maeda, T. et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science 316, 860–866 (2007).
Tan, J. B., Visan, I., Yuan, J. S. & Guidos, C. J. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nature Immunol. 6, 671–679 (2005).
Krueger, A. & von Boehmer, H. Identification of a T lineage-committed progenitor in adult blood. Immunity 26, 105–116 (2007).
Laurent, J., Bosco, N., Marche, P. N. & Ceredig, R. New insights into the proliferation and differentiation of early mouse thymocytes. Int. Immunol. 16, 1069–1080 (2004).
Prockop, S. E. & Petrie, H. T. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J. Immunol. 173, 1604–1611 (2004).
Yücel, R., Karsunky, H., Klein-Hitpass, L. & Möröy, T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 197, 831–844 (2003).
Hock, H. et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003).
Schilham, M. W. et al. Critical involvement of Tcf-1 in expansion of thymocytes. J. Immunol. 161, 3984–3991 (1998).
Weerkamp, F. et al. Wnt signaling in the thymus is regulated by differential expression of intracellular signaling molecules. Proc. Natl Acad. Sci. USA 103, 3322–3326 (2006).
Rosenbauer, F. et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nature Genet. 38, 27–37 (2006).
Hager-Theodorides, A. L. et al. Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J. Immunol. 169, 5496–5504 (2002).
Tsai, P. T., Lee, R. A. & Wu, H. BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood 102, 3947–3953 (2003).
Varas, A. et al. The role of morphogens in T-cell development. Trends Immunol. 24, 197–206 (2003).
Tsuji, M., Shinkura, R., Kuroda, K., Yabe, D. & Honjo, T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc. Natl Acad. Sci. USA 104, 1610–1615 (2007). Deficiency of an inhibitor of Notch-dependent transcription reveals that Notch effects on early T cells depend on feedback negative regulation. The action of MINT and effects on target gene Nrarp distinguish Notch promotion of ETP self renewal from Notch promotion of DN2 cell generation.
Yun, T. J. & Bevan, M. J. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170, 5834–5841 (2003).
Laiosa, C. V., Stadtfeld, M. & Graf, T. Determinants of lymphoid-myeloid lineage diversification. Annu. Rev. Immunol. 24, 705–738 (2006).
Laiosa, C. V., Stadtfeld, M., Xie, H., Andres-Aguayo, L. & Graf, T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25, 731–744 (2006). This study shows that re-expression of either C/EBPα or PU.1 in fully-committed DN3 cells is sufficient to reverse commitment and transform the thymocytes to myeloid or DCs. The authors show some rescue of Thy-1+ cells by coexpression of NotchIC or GATA3, but not by E2A.
Dionne, C. J. et al. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev. Biol. 280, 448–466 (2005).
Lefebvre, J. M. et al. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J. Immunol. 174, 6184–6194 (2005).
Boos, M. D., Yokota, Y., Eberl, G. & Kee, B. L. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204, 1119–1130 (2007).
Ikawa, T., Fujimoto, S., Kawamoto, H., Katsura, Y. & Yokota, Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc. Natl Acad. Sci. USA 98, 5164–5169 (2001).
Iwasaki-Arai, J., Iwasaki, H., Miyamoto, T., Watanabe, S. & Akashi, K. Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J. Exp. Med. 197, 1311–1322 (2003).
Hsu, C. L. et al. Antagonistic effect of CCAAT enhancer-binding protein-α and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc. Natl Acad. Sci. USA 103, 672–677 (2006).
Shibata, Y. et al. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity 15, 557–567 (2001).
Carotta, S., Brady, J., Wu, L. & Nutt, S. L. Transient Notch signaling induces NK cell potential in Pax5-deficient pro-B cells. Eur. J. Immunol. 36, 3294–3304 (2006).
Rolink, A. G., Balciunaite, G., Demoliere, C. & Ceredig, R. The potential involvement of Notch signaling in NK cell development. Immunol. Lett. 107, 50–57 (2006).
Yokota, Y. & Mori, S. Role of Id family proteins in growth control. J. Cell. Physiol. 190, 21–28 (2002).
Brunet de la Grange, P. et al. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood 108, 2998–3004 (2006).
Curtis, D. J. et al. SCL is required for normal function of short-term repopulating hematopoietic stem cells. Blood 103, 3342–3348 (2004).
Fisher, R. C., Lovelock, J. D. & Scott, E. W. A critical role for PU.1 in homing and long-term engraftment by hematopoietic stem cells in the bone marrow. Blood 94, 1283–1290 (1999).
Krosl, G. et al. Transcription factor SCL is required for c-kit expression and c-Kit function in hemopoietic cells. J. Exp. Med. 188, 439–450 (1998).
Matushansky, I., Radparvar, F. & Skoultchi, A. I. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene 22, 4143–4149 (2003).
Kawazu, M. et al. Functional domains of Runx1 are differentially required for CD4 repression, TCRβ expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. J. Immunol. 174, 3526–3533 (2005).
Lieu, Y. K., Kumar, A., Pajerowski, A. G., Rogers, T. J. & Reddy, E. P. Requirement of c-myb in T cell development and in mature T cell function. Proc. Natl Acad. Sci. USA 101, 14853–14858 (2004).
Bender, T. P., Kremer, C. S., Kraus, M., Buch, T. & Rajewsky, K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nature Immunol. 5, 721–729 (2004).
Emambokus, N. et al. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 22, 4478–4488 (2003).
Growney, J. D. et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494–504 (2005).
Inoue, J. et al. Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβ T cells in Bcl11b−/− mice. J. Immunol. 176, 5871–5879 (2006).
Ciofani, M. & Zúñiga-Pflücker, J. C. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nature Immunol. 6, 881–888 (2005).
Weng, A. P. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109 (2006).
Palomero, T. et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. USA 103, 18261–18266 (2006).
Barndt, R. J., Dai, M. & Zhuang, Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20, 6677–6685 (2000).
Wojciechowski, J., Lai, A., Kondo, M. & Zhuang, Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J. Immunol. 178, 5717–5726 (2007). Using a double conditional knockout mouse system, the authors show that cells which reach the DN3 stage before deleting E2A and HEB seem to reverse course after deleting them, and return to extended IL-7-dependent proliferation and a DN2-like phenotype.
Schwartz, R., Engel, I., Fallahi-Sichani, M., Petrie, H. T. & Murre, C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl Acad. Sci. USA 103, 9976–9981 (2006).
Engel, I. & Murre, C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 23, 202–211 (2004).
Visan, I. et al. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nature Immunol. 7, 634–643 (2006).
Besseyrias, V. et al. Hierarchy of Notch-Delta interactions promoting T cell lineage commitment and maturation. J. Exp. Med. 204, 331–343 (2007).
Engel, I., Johns, C., Bain, G., Rivera, R. R. & Murre, C. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194, 733–746 (2001).
Jepsen, K. et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102, 753–763 (2000).
Pennington, D. J. et al. The inter-relatedness and interdependence of mouse T cell receptor γδ+ and αβ+ cells. Nature Immunol. 4, 991–998 (2003).
Tanigaki, K. et al. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20, 611–622 (2004).
Xue, H.-H. et al. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nature Immunol. 5, 1036–1044 (2004).
Porritt, H. E. et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20, 735–745 (2004).
Hoffman, E. S. et al. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 10, 948–962 (1996).
Izon, D. J. et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 16, 231–243 (2002).
Okamura, R. M. et al. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8, 11–20 (1998).
Acknowledgements
The authors wish to thank T. Graf, H. Kawamoto, B. Kee, C. Murre, H. Petrie and N. Speck for valuable discussions and generous sharing of unpublished results. We apologize to many authors whose work we could not adequately cite. We also thank members of the Rothenberg group for collegial interchange, advice and permission to cite unpublished work, and N. Feng, R. Butler and D. Perez for technical support. The authors were supported by grants from the National Institutes of Health (NIH), USA, to E.V.R. (R01 CA90233 and R01 CA98925), M.A.Y. (R01 AI064590) and J.E.M. (F32 AI068366). E.V.R. gratefully acknowledges the Albert Billings Ruddock Professorship.
Author information
Authors and Affiliations
Corresponding author
Related links
FURTHER INFORMATION
Glossary
- T-cell receptor (TCR) gene rearrangement
-
A process whereby the clusters of interchangeable DNA segments that encode TCR genes are recombined to assemble highly diverse TCR structures. High structural diversity is obtained as a result of combinatorial joining and mutations at the joining sites. All TCR gene rearrangement is mediated by RAG1 (recombination-activating gene 1)–RAG2 complexes.
- NKT cells
-
A heterogeneous subset of natural-killer αβ T cells characterized by their expression of semi-invariant T-cell receptor α-chains together with NK-cell markers, and which are positively selected by interaction with non-classical MHC class I molecules. They perform regulatory and effector functions at the interface of innate and adaptive immune responses.
- Regulatory T cells
-
(TReg cells). A rare population of CD4+ T cells that naturally express high levels of CD25 and the transcription factor forkhead box P3 (FOXP3), and that have suppressive regulatory activity towards effector T cells and other immune cells in the periphery. Many of them are thought to be generated by a special intrathymic alternative to negative selection.
- KIT
-
A receptor protein tyrosine kinase, expressed by melanocytes, germ cells, stem cells and most immature haematopoietic cells. KIT binds the growth factor stem-cell factor (SCF), also known as KIT ligand or Steel factor.
- β-selection
-
The controlled developmental transition beyond the double-negative 3 (DN3) stage to the double positive (DP) stage that is limited to T cells that have successfully rearranged their T-cell receptor (TCR) β-chain genes to express a functional cell-surface pre-TCR. The conditional developmental arrest encountered at the DN3 stage is termed the 'β-selection checkpoint'.
- Positive selection
-
A step in the process of T-cell differentiation in the thymus that selects CD4+CD8+ double-positive T cells for survival and maturation, based on the appropriate degree of interaction between their T-cell receptor and peptide–MHC complexes on thymic epithelial cells. Depending on the class of MHC molecule recognized, thymocytes are positively selected either to a CD4+ or to a CD8+ single-positive cell fate.
- Negative selection
-
The intrathymic elimination of double-positive or single-positive thymocytes that express T-cell receptors with high affinity for self antigens.
- Notch
-
A transmembrane receptor involved in the pathway for direct cell–cell signalling through its association with a transmembrane ligand of the delta or serrate/jagged family on a neighbouring cell. The large intracellular domain of Notch is cleaved and travels to the nucleus to become a direct co-activator of the transcription factor RBPJ (CSL).
- Interleukin-7
-
(IL-7). The major growth factor for immature T and B cells and for homeostatic proliferation of post-thymic naive T cells. It triggers a heterodimeric receptor composed of the IL-7 receptor α-chain (also known as CD127) coupled with the common γ chain (also known as CD132).
- Recombination activating gene 1 (RAG1) and RAG2
-
A recombinase complex encoded by a pair of linked genes. RAG1–RAG2 complexes are essential for all V(D)J-type rearrangement of immunoglobulin genes as well as for all TCR gene rearrangements.
- HEBalt
-
An alternative short form of the transcription factor HEB (HeLa E-box-binding protein; also known as TCF12), which has the same DNA binding and dimerization domains as the canonical form of HEB but which has an alternative N-terminus and distinct functionality.
- Hedgehog signalling pathway
-
A signalling pathway that promotes growth or differentiation during embryonic development and T-cell development. Hedgehog family soluble ligands activate transcription factors of the GLI (glioma-associated oncogene) family through a complex pathway involving two transmembrane receptors, Patched and Smoothened, and a double inhibition system.
- WNT
-
A signalling mediator named both for its mutant phenotype in Drosophila melanogaster (Wingless) and for its role as a preferential retrovirus integration site in murine leukaemia virus-induced leukaemias (Int-1). WNT signalling activates the TCF1 and LEF1 family transcription factors through stabilizing their co-activator, β-catenin, and mobilizing it from the cytoplasm to the nucleus.
- HEBcan
-
Designation for the canonical form of HEB (HeLa E-box-binding protein; also known as TCF12), as contrasted with HEBalt.
Rights and permissions
About this article
Cite this article
Rothenberg, E., Moore, J. & Yui, M. Launching the T-cell-lineage developmental programme. Nat Rev Immunol 8, 9–21 (2008). https://doi.org/10.1038/nri2232
Issue Date:
DOI: https://doi.org/10.1038/nri2232
This article is cited by
-
Epigenetic reprogramming of T cells: unlocking new avenues for cancer immunotherapy
Cancer and Metastasis Reviews (2024)
-
Generating hematopoietic cells from human pluripotent stem cells: approaches, progress and challenges
Cell Regeneration (2023)
-
TAL1 hijacks MYCN enhancer that induces MYCN expression and dependence on mevalonate pathway in T-cell acute lymphoblastic leukemia
Leukemia (2023)
-
Chemokine receptor CCR9 suppresses the differentiation of CD4+CD8αα+ intraepithelial T cells in the gut
Mucosal Immunology (2022)
-
TCF-1: a maverick in T cell development and function
Nature Immunology (2022)