Key Points
-
Speckles are dynamic subnuclear structures that contain pre-messenger RNA splicing factors and other proteins that are involved in transcription, 3′- end RNA-processing and reversible protein phosphorylation. The formation of speckles is regulated during the cell-division cycle.
-
Splicing factors cycle continually between speckles and the nucleoplasm. Their size and shape results from the dynamic exchange of factors into and out of speckles.
-
A reversible protein phosphorylation mechanism can regulate the movement of speckle components between speckles and other nuclear structures. It is likely that protein–protein interactions are primarily responsible for the formation and integrity of speckles.
-
Speckles contain little or no DNA and are not principal sites of transcription. Instead, they function as assembly/modification sites that can supply active splicing factors to sites of transcription.
-
We propose a 'regulated-exchange' model to account for the steady-state level of proteins in speckles. This envisages that the concentration of factors that are localized in speckles results from a regulated and cell-type-specific basal exchange rate of speckle components.
Abstract
Speckles are subnuclear structures that are enriched in pre-messenger RNA splicing factors and are located in the interchromatin regions of the nucleoplasm of mammalian cells. At the fluorescence-microscope level they appear as irregular, punctate structures, which vary in size and shape, and when examined by electron microscopy they are seen as clusters of interchromatin granules. Speckles are dynamic structures, and both their protein and RNA–protein components can cycle continuously between speckles and other nuclear locations, including active transcription sites. Studies on the composition, structure and behaviour of speckles have provided a model for understanding the functional compartmentalization of the nucleus and the organization of the gene-expression machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Misteli, T. Protein dynamics: implications for nuclear architecture and gene expression. Science 291, 843–847 (2001).
Spector, D. L. Nuclear bodies. J. Cell Sci. 114, 2891–2893 (2001).
Lamond, A. I. & Earnshaw, W. C. Structure and function in the nucleus. Science 280, 547–553 (1998).
Wansink, D. G. et al. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122, 283–293 (1993).
Phair, R. D. & Misteli, T. High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609 (2000). Photobleaching techniques show that many classes of proteins can move rapidly within the nucleus and that they can rapidly associate and dissociate with nuclear compartments.
Beck, J. S. Variations in the morphological patterns of "autoimmune" nuclear fluorescence. Lancet 1, 1203–1205 (1961).
Swift, H. Studies on nuclear fine structure. Brookhaven Symp. Biol. 12, 134–152 (1959).
Perraud, M., Gioud, M. & Monier, J. C. Intranuclear structures of monkey kidney cells recognised by immunofluorescence and immuno-electron microscopy using anti-ribonucleoprotein antibodies. Ann. Immunol. 130, 635–647 (1979) (in French).
Lerner, E. A., Lerner, M. R., Janeway, C. A. & Steitz, J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA 78, 2737–2741 (1981).
Spector, D. L., Schrier, W. H. & Busch, H. Immunoelectron microscopic localization of snRNPs Biol. Cell 49, 1–10 (1983).
Thiry, M. The interchromatin granules. Histol. Histopathol. 10, 1035–1045 (1995).
Wei, X., Somanathan, S., Samarabandu, J. & Berezney, R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 146, 543–558 (1999).
Visa, N., Puvion-Dutilleul, F., Bachellerie, J. P. & Puvion, E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur. J. Cell Biol. 60, 308–321 (1993).
Spector, D. L. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265–315 (1993).
Fakan, S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 4, 86–90 (1994).
Li, H. & Bingham, P. M. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67, 335–342 (1991).
Hedley, M. L., Amrein, H. & Maniatis, T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine rich splicing factor. Proc. Natl Acad. Sci. USA 92, 11524–11528 (1995).
Caceres, J. F., Misteli, T., Screaton, G. R., Spector, D. L. & Krainer, A. R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138, 225–238 (1997).
Gall, J. G., Bellini, M., Wu, Z. & Murphy, C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10, 4385–4402 (1999).
Segalat, L. & Lepesant, J. A. Spatial distribution of the Sm antigen in Drosophila early embryos. Biol. Cell 75, 181–185 (1992).
Potashkin, J. A., Derby, R. J. & Spector, D. L. Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Mol. Cell. Biochem. 10, 3524–3534 (1990).
Fox, A. H. et al. Paraspeckles: a novel nuclear domain. Curr. Biol. 12, 13–25 (2002).
Huang, S. & Spector, D. L. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 5, 2288–2302 (1991).
Xing, Y., Johnson, C. V., Dobner, P. R. & Lawrence, J. B. Higher level organization of individual gene transcription and RNA splicing. Science 259, 1326–1330 (1993).
Xing, Y., Johnson, C. V., Moen, P. T., McNeil, J. A. & Lawrence, J. B. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC35 domains. J. Cell Biol. 131, 1635–1647 (1995).
Smith, K. P., Moen, P. T., Wydner, K. L., Coleman, J. R. & Lawrence, J. B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144, 617–629 (1999).
Johnson, C. et al. Tracking COL1A1 RNA in osteogenesis imperfecta. Splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 150, 417–432 (2000).
Misteli, T., Cáceres, J. F. & Spector, D. L. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387, 523–527 (1997). The first study to show that speckles are dynamic structures that respond to activation of nearby genes, by using live imaging of cells expressing a fluorescent-protein-tagged splicing factor.
O'Keefe, R. T., Mayeda, A., Sadowski, C. L., Krainer, A. R. & Spector, D. L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249–260 (1994).
Monneron, A. & Bernhard, W. Fine structural organization of the interphase nucleus in some mammalian cells. J. Ultrastruct. Res. 27, 266–288 (1969).
Fakan, S. & Bernhard, W. Localisation of rapidly and slowly labelled nuclear RNA as visualized by high resolution autoradiography. Exp. Cell Res. 67, 129–141 (1971).
Fakan, S. & Nobis, P. Ultrastructural localization of transcription sites and of RNA distribution during the cell cycle of synchronized CHO cells. Exp. Cell Res. 113, 327–337 (1978).
Cmarko, D. et al. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 10, 211–223 (1999). This study showed that transcription is associated with perichromatin fibrils and not with nuclear speckles (IGCs).
Huang, S. & Spector, D. L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 131, 719–732 (1996).
Melcak, I. et al. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell 11, 497–510 (2000).
Shopland, L. S., Johnson, C. V. & Lawrence, J. B. Evidence that all SC35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J. Struct. Biol. 140, 131–139 (2002).
Fu, X. -D. The superfamily of arginine/serine-rich splicing factors. RNA 1, 663–680 (1995).
Huang, S. & Spector, D. L. in Eukaryotic mRNA Processing (ed. Krainer, A. R.) 37–67 (Oxford Univ. Press, New York, 1997).
Colwill, K. et al. The Clk/Sty protein kinase phosphorylates splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265–275 (1996).
Ko, T. K., Kelly, E. & Pines, J. CrkRS: a novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 114, 2591–2603 (2001).
Kojima, T., Zama, T., Wada, K., Onogi, H. & Hagiwara, M. Cloning of human PRP4 reveals interaction with Clk1. J. Biol. Chem. 276, 32247–32256 (2001).
Sacco-Bubulya, P. & Spector, D. L. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156, 425–436 (2002).
Brede, G., Solheim, J. & Prydz, H. PSKH1, a novel splice factor compartment-associated serine kinase. Nucleic Acids Res. 30, 5301–5309 (2002).
Trinkle-Mulcahy, L. et al. Nuclear organisation of NIPP1, a regulatory subunit of protein phosphatase 1 that associates with pre-mRNA splicing factors. J. Cell Sci. 112, 157–168 (1999).
Trinkle-Mulcahy, L., Sleeman, J. E. & Lamond, A. I. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 114, 4219–4228 (2001).
Misteli, T. & Spector, D. L. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 7, 135–138 (1997).
Mintz, P. J., Patterson, S. D., Neuwald, A. F., Spahr, C. S. & Spector, D. L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18, 4308–4320 (1999). The first systematic proteomic study of the protein components of nuclear speckles (IGCs), which were isolated from mouse liver nuclei.
Larsson, S. H. et al. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 81, 391–401 (1995).
Mortillaro, M. J. et al. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl Acad. Sci. USA 93, 8253–8257 (1996).
Zeng, C., Kim, E., Warren, S. L. & Berget, S. M. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16, 1401–1412 (1997).
Krause, S., Fakan, S., Weis, K. & Wahle, E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 214, 75–82 (1994).
Schul, W., van Driel, R. & de Jong, L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp. Cell Res. 238, 1–12 (1998).
Dostie, J., Lejbkowicz, F. & Sonenberg, N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol. 148, 239–247 (2000).
Li, Q. et al. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 19, 7336–7346 (1999).
Nakayasu, H. & Ueda, K. Small nuclear RNA–protein complex anchors on the actin filaments in bovine lymphocyte nuclear matrix. Cell Struct. Funct. 9, 317–325 (1984).
Jagatheesan, G. et al. Colocalization of intranuclear lamin foci with RNA splicing factors. J. Cell Sci. 112, 4651–4661 (1999).
Rappsilber, J., Ryder, U., Lamond, A. I. & Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245 (2002).
Zhou, Z., Licklider, L. J., Gygi, S. P. & Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185 (2002).
Bregman, D. B., Du, L., van der Zee, S. & Warren, S. L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129, 287–298 (1995).
Grande, M. A., van der Kraan, I., de Jong, L. & van Driel, R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 110, 1781–1791 (1997).
Kimura, H., Sugaya, K. & Cook, P. R. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159, 777–782 (2002).
Doyle, O., Corden, J. L., Murphy, C. & Gall, J. G. The distribution of RNA polymerase II largest subunit (RPB1) in the Xenopus germinal vesicle. J. Struct. Biol. 140, 154–166 (2002).
Price, D. H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20, 2629–2634 (2000).
Herrmann, C. H. & Mancini, M. A. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114, 1491–1503 (2001).
Matera, A. G. & Ward, D. C. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 121, 715–727 (1993).
Pessler, F., Pendergrast, P. S. & Hernandez, N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol. Cell. Biol. 17, 3786–3798 (1997).
Pendergrast, P. S., Wang, C., Hernandez, N. & Huang, S. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-P-TEFb-containing nuclear speckles. Mol. Biol. Cell 13, 915–929 (2002).
Hock, R., Wilde, F., Scheer, U. & Bustin, M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 17, 6992–7001 (1998).
Carter, K. C., Taneja, K. L. & Lawrence, J. B. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 115, 1191–1202 (1991).
Visa, N., Puvion-Dutilleul, F., Harper, F., Bachellerie, J. -P. & Puvion, E. Intranuclear distribution of poly A RNA determined by electron microscope in situ hybridization. Exp. Cell. Res. 208, 19–34 (1993).
Huang, S., Deerinck, M. H., Ellisman, M. H. & Spector, D. L. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 126, 877–899 (1994).
Erdmann, V. A., Szymanski, M., Hochberg, A., de Groot, N. & Barciszewski, J. Collection of mRNA-like non-coding RNAs. Nucleic Acids Res. 27, 192–195 (1999).
Herman, R. C., Williams, J. G. & Penman, S. Message and non-message sequences adjacent to poly(A) in steady state heterogeneous nuclear RNA of HeLa cells Cell 7, 429–437 (1976).
Zhao, K. et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 (1998).
Boronenkov, I. V., Loijens, J. C., Umeda, M. & Anderson, R. A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 9, 3547–3560 (1998).
Spann, T. P., Goldman, A. E., Wang, C., Huang, S. & Goldman, R. D. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156, 603–608 (2002).
Spector, D. L., Fu, X. -D. & Maniatis, T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10, 3467–3481 (1991).
Jiménez-García, L. F. & Spector, D. L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73, 47–59 (1993). This work showed that factors are recruited from nuclear speckles to sites of transcription.
Bridge, E. et al. Dynamic organization of splicing factors in adenovirus-infected cells. J. Virol. 69, 281–290 (1995).
Kruhlak, M. J. et al. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 150, 41–51 (2000). This study showed that the dynamics of splicing factor SF2/ASF are consistent with frequent but transient interactions with relatively immobile nuclear binding sites.
Spector, D. L. & Smith, H. C. Redistribution of U-snRNPs during mitosis. Exp. Cell Res. 163, 87–94 (1986).
Reuter, R., Appel, B., Rinke, J. & Lührmann, R. Localization and structure of snRNPs during mitosis. Immunofluorescent and biochemical studies. Exp. Cell Res. 159, 63–79 (1985).
Ferreira, J. A., Carmo-Fonseca, M. & Lamond, A. I. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J. Cell Biol. 126, 11–23 (1994). This study identified MIGs as mitotic forms of speckles containing splicing factors and showed that interphase speckles can reform after mitosis, even in the absence of transcription.
Thiry, M. Behavior of interchromatin granules during the cell cycle Eur. J. Cell Biol. 68, 14–24 (1995). Analysis of the organization of IGCs through the cell cycle at the electron-microscope level.
Leser, G. P., Fakan, S. & Martin, T. E. Ultrastructural distribution of ribonucleoprotein complexes duirng mitosis. snRNP antigens are contained in mitotic granule clusters. Eur. J. Cell Biol. 50, 376–389 (1989).
Verheijen, R., Kuijpers, H., Vooijs, P., Van Venrooij, W. & Ramaekers, F. Distribution of the 70K U1 RNA-associated protein during interphase and mitosis. Correlation with other U RNP particles and proteins of the nuclear matrix. J. Cell Sci. 86, 173–190 (1986).
Prasanth, K. V., Sacco-Bubulya, P., Prasanth, S. G. & Spector, D. L. Sequential entry of components of gene expression machinery into daughter nuclei. Mol. Biol. Cell 14, 1043–1057 (2003). This study shows the differential timing of the re-entry of separate components of the gene-expression machinery into newly assembled daughter nuclei after mitosis. This re-entry occurs in a sequential and ordered manner.
Thiry, M. Differential location of nucleic acids within interchromatin granule clusters. Eur. J. Cell Biol. 62, 259–269 (1993).
Misteli, T. & Spector, D. L. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell 7, 1559–1572 (1996).
Spector, D. L., Lark, G. & Huang, S. Differences in snRNP localization between transformed and nontransformed cells. Mol. Biol. Cell 3, 555–569 (1992).
Spector, D. L., O'Keefe, R. T. & Jiménez-García, L. F. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harb. Symp. Quant. Biol. 58, 799–805 (1993).
Gui, J. F., Lane, W. S. & Fu, X. -D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369, 678–682 (1994). Identifies and characterizes the role of a serine kinase, SRPK1, which phosphorylates SR proteins and modulates their localization during the cell cycle.
Gui, J. F., Tronchere, H., Chandler, S. D. & Fu, S. D. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl Acad. Sci. USA 91, 10824–10828 (1994).
Misteli, T. et al. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143, 297–307 (1998).
Mermoud, J. E., Cohen, P. T. W. & Lamond, A. I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 13, 5679–5688 (1994).
Turner, B. M. & Franchi, L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J. Cell Sci. 87, 269–282 (1987).
Pollard, T. D. & Earnshaw, W. C. Cell Biology (Saunders, Philadelphia, 2002).
Acknowledgements
We thank members of the Lamond group, and K.V. Prasanth and P. Sacco-Bubulya in the Spector group for their helpful comments. We also appreciate the comments of S. Fakan, J. Gall, G. Matera and J. Swedlow. We thank A. Fox and Y. Wah Lam (University of Dundee, UK) for their help in preparing figures 1 and 3, and P. Sacco-Bubulya (Cold Spring Harbor Laboratory) for providing figure 4. A. I. L. is a Wellcome Trust Principal Research Fellow. D. L. S. is funded by the National Institute of General Medical Sciences/ National Institutes of Health.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Interpro
Locus Link
Swiss-Prot
FURTHER INFORMATION
Glossary
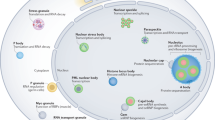
- SPECKLE
-
An irregularly shaped nuclear domain that is visualized by immunofluorescence microscopy, typically by using anti-splicing-factor antibodies. Usually 25–50 speckles are observed per interphase mammalian nucleus.
- PARASPECKLE
-
A subnuclear structure that is distinct from speckles. Typically, 10–20 paraspeckles are present in the interchromatin nucleoplasmic space, and they are often located adjacent to speckles. So far, three proteins — paraspeckle proteins 1 and 2 and p54/nrb — have been localized to these nuclear domains.
- CAJAL BODY
-
A nuclear structure that contains newly assembled small nuclear ribonucleoprotein particles that are involved in pre-messenger RNA splicing, and small nucleolar ribonucleoprotein particles that are involved in ribosomal RNA processing. Also contains Cajal-body-specific guide RNAs. Cajal bodies are usually identified as foci labelled with antibodies against the autoantigen p80 coilin.
- GEMS
-
'Gemini of Cajal bodies' are nuclear structures that are usually localized either coincident with or adjacent to Cajal bodies, depending on the cell line examined. Gems are characterized by the presence of the 'survival of motor neurons' (SMN) protein.
- PROMYELOCYTIC LEUKAEMIA (PML) BODY
-
A subnuclear structure that is also known as nuclear domain 10, promyelocytic leukaemia oncogenic domain or Kr body. These bodies are characterized by the presence of the promyelocytic leukaemia protein and there are typically 10–30 per nucleus.
- INTERCHROMATIN GRANULE CLUSTER
-
(IGC). A structure seen by electron microscopy that is equivalent to the speckles that are seen by fluorescence microscopy. Each IGC is composed of a series of particles, 20–25 nm in diameter, that seem to be connected in places by a thin fibril.
- PERICHROMATIN FIBRILS
-
Fibrils observed by the electron microscope that are detected at transcription sites and shown to coincide with the incorporation of tritiated-uridine or 5-bromouridine 5′-trisphosphate, indicating that they are nascent transcripts.
- INTERCHROMATIN-GRANULE-ASSOCIATED ZONE
-
A region that is adjacent to interchromatin granule clusters, which contains U1, but not U2, small nuclear RNAs.
- SR PROTEINS
-
A family of pre-messenger RNA splicing factors that are characterized by repeats of arginine–serine dipeptides at their carboxyl termini.
- SNURPOSOME
-
A nuclear structure, identified in amphibian oocytes, that contains splicing small nuclear ribonucleoprotein particles. Three classes — known as A, B and C snurposomes — have been defined, and they differ in their composition. B snurposomes are most closely related to speckles in their composition, and could represent oocyte forms of the speckles that are found in somatic-cell nuclei.
- CELLULARIZATION
-
The transition from a syncytium to distinct cells which occurs at the fourteenth round of cell division in the Drosophila melanogaster embryo.
- CLK/STY
-
A kinase family, the members of which are characterized by having the serine residues in the arginine–serine domain of SR proteins as their primary substrates.
- PRP4
-
A kinase that localizes to nuclear speckles and interacts with CLK/STY, as well as several proteins that are involved in pre-mRNA splicing (SF2/ASF, U5 snRNP) and chromatin remodelling (BRG1, N-CoR deacetylase complexes).
- PSKH1
-
A human kinase that is localized to nuclear speckles but that does not directly interact with SR proteins.
- MITOTIC INTERCHROMATIN GRANULES
-
(MIGS). The speckles (interchromatin granule clusters) that form in the cytoplasm of cells undergoing mitosis, and that increase in number from metaphase to telophase.
- REGULATED-EXCHANGE MODEL
-
A model proposed in this review to account for the basic principles of speckle formation and their dynamic properties.
Rights and permissions
About this article
Cite this article
Lamond, A., Spector, D. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4, 605–612 (2003). https://doi.org/10.1038/nrm1172
Issue Date:
DOI: https://doi.org/10.1038/nrm1172
This article is cited by
-
A model for organization and regulation of nuclear condensates by gene activity
Nature Communications (2023)
-
Formation of nuclear CPSF6/CPSF5 biomolecular condensates upon HIV-1 entry into the nucleus is important for productive infection
Scientific Reports (2023)
-
RNA-mediated demixing transition of low-density condensates
Nature Communications (2023)
-
Nuclear speckleopathies: developmental disorders caused by variants in genes encoding nuclear speckle proteins
Human Genetics (2023)
-
From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo
Nature Reviews Genetics (2022)