Key Points
-
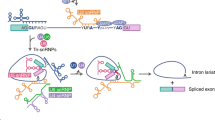
Unlike transcription, where regulation is initiated most frequently by high-affinity, sequence-specific DNA-binding proteins, splicing regulation is frequently brought about by cooperative interactions between factors such as serine-arginine rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), which have lower affinities and sequence specificities.
-
HnRNP A1 represses variant exon-5 (v5) inclusion in human CD44 pre-mRNA in most tissues. In T cells that are activated by Ras signalling, SAM68, which is phosphorylated by extracellular signal-regulated kinase (ERK), interacts with v5 RNA in a way that interferes with the repressive activity of hnRNP A1, and enhances v5 inclusion in the final CD44 mRNA.
-
Fas-mediated signalling regulates the alternative splicing of transcripts that encode apoptotic regulators. Alterations in the phosphorylation status of SR proteins probably lead to changes in alternative splicing of BCL-X pre-mRNAs. Additionally, Fas-mediated phosphorylation of TIA1 might modulate the alternative splicing of Fas pre-mRNA, thereby providing a positive autoregulatory loop to enhance Fas signalling.
-
SRp38 functions as a splicing repressor when it is activated by dephosphorylation during M phase of the cell cycle and in response to heat shock. Splicing inhibition is brought about by an interaction between dephosphorylated SRp38, U1 small nuclear ribonucleoprotein particles (U1 snRNPs), and possibly other snRNPs.
-
Ania-6–cyclin-dependent-kinasep100 (CDK11p100) provides a link between dopamine signalling and transcription-coupled splicing by phosphorylating SR proteins and the carboxy-terminal domain of the largest subunit of RNA polymerase II (RNAPII CTD).
-
After depolarization of GH3 pituitary cells, the inclusion of the stress-axis-regulated (STREX) exon of the SLO pre-mRNA is repressed by the CaRRE (Ca2+/calmodulin-dependent protein kinase (CaMK)IV-responsive RNA element)-specific binding of an unidentified protein that is phosphorylated by CaMKIV.
Abstract
The transcripts of most metazoan protein-coding genes are alternatively spliced, but the mechanisms that are involved in the control of splicing are not well understood. Recent evidence supports the potential of both extra- and intracellular signalling to the splicing machinery as a means of regulating gene expression, and indicates that this form of gene control is widespread and mechanistically complex. However, important questions about these pathways need to be answered before this method of post-transcriptional regulation can be fully appreciated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Mironov, A. A., Fickett, J. W. & Gelfand, M. S. Frequent alternative splicing of human genes. Genome Res. 9, 1288–1293 (1999).
Johnson, J. M. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 (2003).
Kan, Z., Rouchka, E. C., Gish, W. R. & States, D. J. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11, 889–900 (2001).
Modrek, B., Resch, A., Grasso, C. & Lee, C. Genome-wide analysis of alternative splicing using human expressed sequence data. Nucleic Acids Res. 29, 2850–2859 (2001).
Blencowe, B. J. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25, 106–110 (2000).
Smith, C. W. & Valcarcel, J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388 (2000).
Grabowski, P. J. & Black, D. L. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65, 289–308 (2001).
Graveley, B. R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17, 100–107 (2001).
Maniatis, T. & Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418, 236–243 (2002).
Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 (2003).
Manley, J. L. & Tacke, R. SR proteins and splicing control. Genes Dev. 10, 1569–1579 (1996).
Graveley, B. R. Sorting out the complexity of SR protein functions. RNA 6, 1197–1211 (2000).
Wu, J. Y. & Maniatis, T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 (1993).
Kohtz, J. D. et al. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124 (1994).
Xiao, S. H. & Manley, J. L. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344 (1997).
Xiao, S. H. & Manley, J. L. Phosphorylation–dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 17, 6359–6367 (1998).
Gui, J. F., Lane, W. S. & Fu, X. D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369, 678–682 (1994).
Colwill, K. et al. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265–275 (1996).
Duncan, P. I. et al. Alternative splicing of STY, a nuclear dual specificity kinase. J. Biol. Chem. 270, 21524–21531 (1995).
Lee, K., Du, C., Horn, M. & Rabinow, L. Activity and autophosphorylation of LAMMER protein kinases. J. Biol. Chem. 271, 27299–27303 (1996).
Nayler, O., Stamm, S. & Ullrich, A. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem. J. 326, 693–700 (1997).
Prasad, J., Colwill, K., Pawson, T. & Manley, J. L. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 19, 6991–7000 (1999).
Prasad, J. & Manley, J. L. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol. Cell. Biol. 23, 4139–4149 (2003).
Ko, T. K., Kelly, E. & Pines, J. CrkRS: a novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 114, 2591–2603 (2001).
Hirose, Y. & Manley, J. L. RNA polymerase II and the integration of nuclear events. Genes Dev. 14, 1415–1429 (2000).
Proudfoot, N. J., Furger, A. & Dye, M. J. Integrating mRNA processing with transcription. Cell 108, 501–512 (2002).
Maniatis, T. & Reed, R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002).
Dreyfuss, G., Matunis, M. J., Pinol-Roma, S. & Burd, C. G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62, 289–321 (1993).
Mayeda, A. & Krainer, A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68, 365–375 (1992).
Singh, R., Valcarcel, J. & Green, M. R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173–1176 (1995).
Ruskin, B., Zamore, P. D. & Green, M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52, 207–219 (1988).
Labourier, E. et al. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 13, 740–753 (1999).
Shin, C. & Manley, J. L. The SR protein SRp38 represses splicing in M phase cells. Cell 111, 407–417 (2002). Characterizes SRp38 as a general splicing repressor that is activated by dephosphorylation, and shows that the splicing machinery is repressed by SRp38 during M phase of the cell cycle.
Kanopka, A., Muhlemann, O. & Akusjarvi, G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381, 535–538 (1996).
Valcarcel, J., Singh, R., Zamore, P. D. & Green, M. R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-RNA. Nature 362, 171–175 (1993).
Amrein, H., Hedley, M. L. & Maniatis, T. The role of specific protein–RNA and protein–protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell 76, 735–746 (1994).
Jensen, K. B. et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359–371 (2000). Describes NOVA1 as a brain-specific splicing regulator by showing neuronal splicing defects in Nova1 -null mice.
Ponta, H., Sherman, L. & Herrlich, P. A. CD44: from adhesion molecules to signaling regulators. Nature Rev. Mol. Cell Biol. 4, 33–45 (2003).
Chang, L. & Karin, M. Mammalian MAP kinase signalling cascades. Nature 410, 37–40 (2001).
Konig, H., Ponta, H. & Herrlich, P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 17, 2904–2913 (1998). Provides an initial characterization of CD44 v5 alternative-splicing regulation in response to activated Ras by identifying positively and negatively acting sequences.
Weg-Remers, S., Ponta, H., Herrlich, P. & Konig, H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 20, 4194–4203 (2001).
Vernet, C. & Artzt, K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13, 479–484 (1997).
Taylor, S. J. & Shalloway, D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 368, 867–871 (1994).
Fumagalli, S., Totty, N. F., Hsuan, J. J. & Courtneidge, S. A. A target for Src in mitosis. Nature 368, 871–874 (1994).
Taylor, S. J., Resnick, R. J. & Shalloway, D. SAM68 exerts separable effects on cell cycle progression and apoptosis. BMC Cell Biol. 5, 5 (2004).
Matter, N., Herrlich, P. & Konig, H. Signal-dependent regulation of splicing via phosphorylation of SAM68. Nature 420, 691–695 (2002). Provides evidence that SAM68 activates CD44 v5 inclusion in response to phosphorylation by ERK.
Tacke, R. & Manley, J. L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14, 3540–3551 (1995).
Tacke, R. Tohyama, M., Ogawa, S., & Manley, J. L. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93, 139–148 (1998).
Matter, N. et al. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 275, 35353–35360 (2000).
Kashima, T. & Manley, J. L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature. Genet. 34, 460–463 (2003).
Hermiston, M. L., Xu, Z. & Weiss, A. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21, 107–137 (2003).
Lynch, K. W. & Weiss, A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol. Cell. Biol. 20, 70–80 (2000).
Lynch, K. W. & Weiss, A. A CD45 polymorphism associated with multiple sclerosis disrupts an exonic splicing silencer. J. Biol. Chem. 276, 24341–24347 (2001).
Rothrock, C., Cannon, B., Hahm, B. & Lynch. K. W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell 12, 1317–1324 (2003). Defines an ESS element as a signal-responsive cis -element that mediates CD45 v5 skipping.
Lemaire, R., Winne, A., Sarkissian, M. & Lafyatis, R. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur. J. Immunol. 29, 823–837 (1999).
ten Dam, G. B. et al. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J. Immunol. 164, 5287–5295 (2000).
Wu, J. Y., Tang, H. & Havlioglu, N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog. Mol. Subcell. Biol. 31, 153–185 (2003).
Chalfant, C. E. et al. FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phasphatase 1. J. Biol. Chem. 276, 44848–44855 (2001).
Bose, R. et al. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell 82, 405–414 (1995).
Utz, P. J., Hottelet, M., van Venrooij, W. J. & Anderson, P. Association of phosphorylated serine/Arg (SR) splicing factors with the U1-small ribonucleoprotein (snRNP) auroantigen complex accompanies apoptotic cell death. J. Exp. Med. 187, 547–560 (1998).
Chalfant, C. E. et al. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J. Biol. Chem. 277, 12587–12595 (2002). Provides evidence that ceramide regulates alternative splicing of transcripts that encode apoptotic regulators.
Massiello, A. et al. Identification of two RNA cis-elements that function to regulate the 5′ splice site selection of Bcl-x pre-mRNA in response to ceramide. J. Biol. Chem. 279, 15799–15804 (2004).
Forch, P. et al. The apoptosis-promoting factor TIA1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6, 1089–1098 (2000). Describes the mechanism by which TIA1 regulates alternative splicing of Fas pre-mRNA.
Forch, P., Puig, O., Martinez, C., Seraphin, B. & Valcarcel, J. The splicing regulator TIA1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21, 6882–6892 (2002).
Tian, Q., Streuli, M., Schlossman, S. F. & Anderson, P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67, 629–639 (1991).
Tian, Q., Taupin, J., Elledge, S., Robertson, M. & Anderson, P. Fas-mediated serine/threonine kinase (FAST) phosphorylates TIA1 during Fas-mediated apoptosis. J. Exp. Med. 182, 865–874 (1995).
Burns, C. G. et al. Removal of a single α-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 801–815 (2002).
Lopez, P. J. & Seraphin, B. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: Implications for splice-site recognition. RNA 5, 1135–1137 (1999).
Segil, N., Guermah, M., Hoffmann, A., Roeder, R. G. & Heintz. N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10, 2389–2400 (1996).
Akoulitchev, S. & Reinberg, D. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12, 3541–3550 (1998).
Long, J. J., Leresche, A., Kriwacki, R. W. & Gottesfeld, J. M. Repression of TFIIH transcriptional activity and TFIIH associated cdk7 kinase activity at mitosis. Mol. Cell. Biol. 18, 1467–1476 (1998).
Xu, Y. X., Hirose, Y., Zhou, X. Z., Lu, K. P. & Manley, J. L. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 17, 2765–2776 (2003).
Colgan, D. F., Murthy, K. G., Prives, C. & Manley, J. L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature 384, 282–285 (1996).
Pyronnet, S., Dostie, J. & Sonenberg, N. Suppression of cap-dependent translation in mitosis. Genes Dev. 15, 2083–2093 (2001).
Shin, C., Feng, Y. & Manley, J. L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558 (2004). Provides evidence that SRp38 is responsible for heat-shock-induced splicing repression in vitro and in vivo.
Pyronnet, S. & Sonenberg, N. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 11, 13–18 (2001).
Trinkle-Mulcahy, L. et al. Nuclear organization of NIPP1, a regulatory subunit of protein phosphotase 1 that associates with pre-mRNA splicing factors. J. Cell Sci. 112, 157–168 (1999).
Beullens, M. & Bollen, M. The protein phosphatase-1 regulator NIPP1 is also a splicing factor involved in a late step of spliceosome assembly. J. Biol. Chem. 277, 19855–19860 (2002).
Boudrez, A. et al. NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J. Biol. Chem. 275, 25411–25417 (2000).
Boudrez, A., Beullens, M., Waelkens, E., Stalmans, W. & Bollen, M. Phosphorylation-dependent interaction between the splicing factors SAP155 and NIPP1. J. Biol. Chem. 277, 31834–31841 (2002).
Anderson, R. A., Boronenkov, I. V., Doughman, S. D., Kunz, J. & Loijens, J. C. Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 274, 9907–9910 (1999).
Toker, A. Phosphoinositides and signal transduction. Cell. Mol. Life Sci. 59, 761–779 (2002).
Boronenkov, I. V., Loijens, J. C., Umeda, M. & Anderson, R. A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 9, 3547–3560 (1998).
Osborne, S. L., Thomas, C. L., Gschmeissner, S. & Schiavo, G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 114, 2501–2511 (2001).
Yost, H. J., Petersen, R. B. & Lindquist, S. RNA metabolism: strategies for regulation in the heat shock response. Trends Genet. 6, 223–227 (1990).
Gattoni, R. et al. The human hnRNP-M proteins: structure and relation with early heat shock-induced splicing arrest and chromosome mapping. Nucleic Acids Res. 24, 2535–2542 (1996).
Mahe, D. et al. Cloning of human 2H9 heterogeneous nuclear ribonucleoproteins. Relation with splicing and early heat shock-induced splicing arrest. J. Biol. Chem. 272, 1827–1836 (1997).
Jenkins, G. M. et al. Acute activation of de novo sphingolipid biosynthesis upon heat shock causes an accumulation of ceramide and subsequent dephosphorylation of SR proteins. J. Biol. Chem. 277, 42572–42578 (2002).
Carlsson, A. A paradigm shift in brain research. Science 294, 1021–1024 (2001).
Nieoullon, A. Dopamine and the regulation of cognition and attention. Prog. Neurobiol. 67, 53–83 (2002).
Berke, J. D., Paletzki, R. F., Aronson, G. J., Hyman, S. E. & Gerfen, C. R. A complex program of striatal gene expression induced by dopaminergic stimulation. J. Neurosci. 18, 5301–5310 (1998).
Berke, J. D. et al. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32, 277–287 (2001).
Sgambato, V., Minassian, R., Nairn, A. C. & Hyman, S. E. Regulation of ania-6 splice variants by distinct signaling pathways in striatal neurons. J. Neurochem. 86, 153–164 (2003).
Boucher, L., Ouzounis, C. A., Enright, A. J. & Blencowe, B. J. A genome-wide survey of RS domain proteins. RNA 7, 1693–1701 (2001).
Dickinson, L. A., Edgar, A. J., Ehley, J. & Gottesfeld, J. M. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J. Biol. Chem. 277, 25465–25473 (2002).
Hu, D., Mayeda, A., Trembley, J. H., Lahti, J. M. & Kidd, V. J. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 278, 8623–8629 (2003).
Trembley, J. H. et al. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem. 277, 2589–2596 (2002).
Lahti, J. M., Xiang, J., Heath, L. S., Campana, D. & Kidd, V. J. PITSLRE protein kinase activity is associated with apoptosis. Mol. Cell. Biol. 15, 1–11 (1995).
Tang, D., Gururajan, R. & Kidd, V. J. Phosphorylation of PITSLRE p110 isoforms accompanies their processing by caspases during Fas-mediated cell death. J. Biol. Chem. 273, 16601–16607 (1998).
de Graaf, K. et al. Characterization of cyclin L2, a novel cyclin with an arginine/serine-rich (RS) domain: phosphorylation by DYRK1A and colocalization with splicing factors. J. Biol. Chem. 279, 4612–4624 (2004).
Yang, L. et al. Cyclin L2, a novel RNA-polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J. Biol. Chem. 279, 11639–11648 (2004). Shows that cyclin L2 functions in splicing in vitro and that overexpression induces apoptosis.
Butler, A., Tsunoda, S., McCobb, D. P., Wei, A. & Salkoff, L. mSlo, a complex mouse gene encoding 'maxi' calcium-activated potassium channels. Science 261, 221–224 (1993).
Black, D. L. Splicing in the inner ear: a familiar tune, but what are the instruments? Neuron 20, 165–168 (1998).
Xie, J. & McCobb, D. P. Control of alternative splicing of potassium channels by stress hormones. Science 280, 443–446 (1998).
Xie, J. & Black, D. L. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature 410, 936–939 (2001). Describes a sequence element in the STREX exon of the SLO transcript that is required for exon exclusion after depolarization-induced signalling through CaMKIV.
Yeakley, J. M. et al. Profiling alternative splicing on fiber-optic arrays. Nature Biotechnol. 20, 353–358 (2002).
Acknowledgements
We are grateful to members of the Manley laboratory for their helpful comments, and to Inna Boluk for help with preparation of the manuscript. Work from the authors' laboratory was supported by grants from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- EXONIC SPLICING ENHANCER
-
(ESE). A sequence that recruits serine-arginine rich (SR) proteins and related factors to pre-mRNA where they promote both constitutive and regulated splicing by interacting with each other, as well as with other factors including snRNP-associated RS-domain-containing proteins. Different ESEs are recognized by specific subsets of SR proteins.
- SRPK
-
The first identified serine-arginine rich (SR)-protein kinase, which phosphorylates Ser residues in the RS domain of SR proteins. The kinase is predominantly localized in the cytoplasm of interphase cells, and shows increased kinase activity during mitosis. In mammals, the SRPK family consists of SRPK1 and SRPK2.
- CLK/STY
-
A member of the CLK/STY-kinase family, which comprises dual-specificity kinases that can potentially phosphorylate Ser/Thr and Tyr residues. Targets include SR proteins, and the kinase itself is extensively autophosphorylated. The mammalian CLK/STY family consists of at least four kinases, CLK/STY, CLK2, CLK3 and CLK4.
- EXONIC SPLICING SILENCER
-
(ESS). A sequence that recruits splicing-repressor proteins such as hnRNP A1. This protein inhibits the formation or stabilization of the snRNP–pre-mRNA complex. Repressor proteins can function directly by binding to SR proteins, or by blocking the binding of SR proteins to downstream ESEs. They can also inhibit the activity of the ESE complex, thereby preventing the use of adjacent 3′ splice sites.
- KH-TYPE RNA-BINDING MOTIF
-
This K-homology (KH) motif, which was originally identified in the human hnRNP K protein, is important for RNA binding and probably binds RNA directly. Several proteins that contain KH-type RNA-binding motifs, including SAM68, are thought to function in signalling pathways, and are members of the signal transduction and activation of RNA (STAR) family.
- CERAMIDE
-
The backbone molecule of sphingolipid, which is a component of membrane lipids. It is generated either by hydrolysis of more complex sphingolipids or by de novo synthesis, and recently has been shown to function as a regulator of the stress response and death-receptor-mediated apoptosis.
- CYCLIN-DEPENDENT KINASE
-
(CDK). A protein kinase that is active only when it is in a complex with a regulatory cyclin protein. CDK–cyclin complexes phosphorylate specific target proteins to trigger different stages of the cell cycle, or different aspects of the RNA polymerase II transcription cycle.
- NUCLEAR SPECKLE
-
An irregularly shaped nuclear organelle that can be visualized by immunofluorescence microscopy using anti-splicing-factor antibodies. Usually, approximately 25–50 speckles are present in the interphase mammalian nucleus, and they are thought to constitute storage/assembly sites for certain splicing factors.
Rights and permissions
About this article
Cite this article
Shin, C., Manley, J. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol 5, 727–738 (2004). https://doi.org/10.1038/nrm1467
Issue Date:
DOI: https://doi.org/10.1038/nrm1467
This article is cited by
-
Stretching muscle cells induces transcriptional and splicing transitions and changes in SR proteins
Communications Biology (2022)
-
Transcriptional response of honey bee (Apis mellifera) to differential nutritional status and Nosema infection
BMC Genomics (2018)
-
ATP7B Mutation Detection and Pathogenicity Analysis: One Atypical Case of Wilson’s Disease with Adrenocortical Insufficiency
Journal of Molecular Neuroscience (2018)
-
SPSB1-mediated HnRNP A1 ubiquitylation regulates alternative splicing and cell migration in EGF signaling
Cell Research (2017)
-
The molecular pathogenesis of chronic lymphocytic leukaemia
Nature Reviews Cancer (2016)