Key Points
-
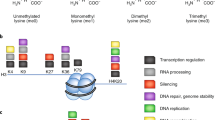
Modification of histone molecules within chromatin has a profound effect on genome structure and function. More specifically, methylation of histone arginine and lysine residues is involved in regulating transcription, epigenetic inheritance and controlling cell fate.
-
Until recently, histone methylation was considered a stable modification. The identification of a histone deiminase and histone demethylases has demonstrated that histone methylation can be dynamically regulated.
-
PADI4 can demethyliminate methylated arginine residues to produce citrulline. Although this reaction does not regenerate arginine, it reveals a mechanism by which arginine methylation can be antagonized.
-
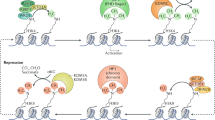
LSD1 uses an amine oxidase reaction to directly remove histone lysine mono- and di-methylation. Removal of H3K4 and H3K9 methylation by LSD1 contributes to transcriptional repression and activation, respectively.
-
JmjC-domain-containing proteins encode a family of histone lysine demethylases that can remove all three methylation states. Members of this family have been shown to catalyse the removal of H3K4, H3K9 and H3K36 methylation.
-
Additional uncharacterized demethylation reaction mechanisms are likely to exist given the extensive complement of methylated histone residues and modification states.
Abstract
Histone methylation has important roles in regulating transcription, genome integrity and epigenetic inheritance. Historically, methylated histone arginine and lysine residues have been considered static modifications because of the low levels of methyl-group turnover in chromatin. The recent identification of enzymes that antagonize or remove histone methylation has changed this view and now the dynamic nature of these modifications is being appreciated. Here, we examine the enzymatic and structural basis for the mechanisms that these enzymes use to counteract histone methylation and provide insights into their substrate specificity and biological function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nature Rev. Mol. Cell Biol. 6, 838–849 (2005).
Lachner, M. & Jenuwein, T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14, 286–298 (2002).
Zhang, Y. & Reinberg, D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 (2001).
Wysocka, J., Allis, C. D. & Coonrod, S. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11, 344–355 (2006).
Wang, H. et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 (2001).
Strahl, B. D. et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11, 996–1000 (2001).
Chen, D. et al. Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 (1999).
Yu, M. C., Lamming, D. W., Eskin, J. A., Sinclair, D. A. & Silver, P. A. The role of protein arginine methylation in the formation of silent chromatin. Genes Dev. 20, 3249–3254 (2006).
Taverna, S. D. et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 (2006).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Huang, Y., Fang, J., Bedford, M. T., Zhang, Y. & Xu, R. M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751 (2006).
Kim, J. et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7, 397–403 (2006).
Wysocka, J. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006).
Shi, X. et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (2006).
Shi, X. et al. Proteome-wide analysis in S. cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455 (2006).
Xiao, B. et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421, 652–656 (2003).
Byvoet, P., Shepherd, G. R., Hardin, J. M. & Noland, B. J. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch. Biochem. Biophys. 148, 558–567 (1972).
Duerre, J. A. & Lee, C. T. In vivo methylation and turnover of rat brain histones. J. Neurochem. 23, 541–547 (1974).
Bannister, A. J. & Kouzarides, T. Reversing histone methylation. Nature 436, 1103–1106 (2005).
Bannister, A. J., Schneider, R. & Kouzarides, T. Histone methylation: dynamic or static? Cell 109, 801–806 (2002).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
Allis, C. D., Bowen, J. K., Abraham, G. N., Glover, C. V. & Gorovsky, M. A. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell 20, 55–64 (1980).
Saccani, S. & Natoli, G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 16, 2219–2224 (2002).
Annunziato, A. T., Eason, M. B. & Perry, C. A. Relationship between methylation and acetylation of arginine-rich histones in cycling and arrested HeLa cells. Biochemistry 34, 2916–2924 (1995).
Nakashima, K., Hagiwara, T. & Yamada, M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 277, 49562–49568 (2002).
Hagiwara, T., Nakashima, K., Hirano, H., Senshu, T. & Yamada, M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem. Biophys. Res. Commun. 290, 979–983 (2002).
Cuthbert, G. L. et al. Histone deimination antagonizes arginine methylation. Cell 118, 545–553 (2004).
Wang, Y. et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279–283 (2004). References 28 and 29 show that PADI4 can antagonize histone arginine methylation by converting methylarginine to citrulline.
Kearney, P. L. et al. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry 44, 10570–10582 (2005).
Arita, K. et al. Structural basis for Ca2+-induced activation of human PAD4. Nature Struct. Mol. Biol. 11, 777–783 (2004).
Lee, Y. H., Coonrod, S. A., Kraus, W. L., Jelinek, M. A. & Stallcup, M. R. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl Acad. Sci. USA 102, 3611–3616 (2005).
Kim, S., Benoiton, L. & Paik, W. K. ε-Alkyllysinase. Purification and properties of the enzyme. J. Biol. Chem. 239, 3790–3796 (1964).
Paik, W. K. & Kim, S. Enzymatic demethylation of calf thymus histones. Biochem. Biophys. Res. Commun. 51, 781–788 (1973). The first demonstration that an enzymatic activity exists in mammalian tissues that can actively demethylate calf thymus histones.
Paik, W. K. & Kim, S. ε-alkyllysinase. New assay method, purification, and biological significance. Arch. Biochem. Biophys. 165, 369–378 (1974).
Hakimi, M. A., Dong, Y., Lane, W. S., Speicher, D. W. & Shiekhattar, R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 278, 7234–7239 (2003).
Humphrey, G. W. et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276, 6817–6824 (2001). Co-REST-containing histone deacetylase complexes are shown to bind FAD and house the amine-oxidase-domain-containing protein LSD1/kiaa0601. The authors speculate that this domain might covalently modify chromatin.
Lee, M. G. et al. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell Biol. 26, 6395–6402 (2006).
Lee, M. G., Wynder, C., Cooch, N. & Shiekhattar, R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435 (2005). References 39 and 44 show that the interaction between LSD1 and Co-REST is important for nucleosomal demethylation.
Shi, Y. et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738 (2003).
Tong, J. K., Hassig, C. A., Schnitzler, G. R., Kingston, R. E. & Schreiber, S. L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 (1998).
You, A., Tong, J. K., Grozinger, C. M. & Schreiber, S. L. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl Acad. Sci. USA 98, 1454–1458 (2001).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). The authors identify the first histone lysine demethylase and demonstrate its role in transcriptional repression.
Shi, Y. J. et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005).
Chen, Y. et al. Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc. Natl Acad. Sci. USA 103, 13956–13961 (2006).
Stavropoulos, P., Blobel, G. & Hoelz, A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nature Struct. Mol. Biol. 13, 626–632 (2006).
Yang, M. et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 23, 377–387 (2006). References 45–47 report the crystal structure of LSD1, and reference 47 also reports on the crystal structure of LSD1–Co-REST, providing insight into the potential mechanisms for nucleosomal demethylation.
Forneris, F., Binda, C., Vanoni, M. A., Battaglioli, E. & Mattevi, A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 280, 41360–41365 (2005).
Ballas, N. et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron 31, 353–365 (2001).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Yamane, K. et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 (2006). Using an unbiased biochemical approach, a JmjC-domain-containing demethylase with specificity toward H3K9 was identified, verifying that the JmjC domain is a histone-demethylase signature motif.
Jarriault, S. & Greenwald, I. Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans. Genes Dev. 16, 2713–2728 (2002).
Eimer, S., Lakowski, B., Donhauser, R. & Baumeister, R. Loss of spr-5 bypasses the requirement for the C. elegans presenilin sel-12 by derepressing hop-1. EMBO J. 21, 5787–5796 (2002).
Dallman, J. E., Allopenna, J., Bassett, A., Travers, A. & Mandel, G. A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J. Neurosci. 24, 7186–7193 (2004).
Nicolas, E. et al. Fission yeast homologs of human histone h3 lysine 4 demethylase regulate a common set of genes with diverse functions. J. Biol. Chem. 281, 35983–35988 (2006).
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T. & Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 (2002).
Falnes, P. O., Johansen, R. F. & Seeberg, E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419, 178–182 (2002).
Trewick, S. C., McLaughlin, P. J. & Allshire, R. C. Methylation: lost in hydroxylation? EMBO Rep. 6, 315–320 (2005). The authors propose that JmjC-domain-containing proteins might function as histone demethylases on the basis of the function of the fission yeast JmjC-domain protein Epe1 in regulating silent chromatin structure.
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). The authors used a novel in vitro histone demethylase assay to biochemically purify and characterize the first JmjC-domain-containing histone demethylase enzyme. This study shows that the JmjC domain is a signature motif for histone demethylases.
Klose, R. J., Kallin, E. M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev. Genet. 7, 715–727 (2006).
Gearhart, M. D., Corcoran, C. M., Wamstad, J. A. & Bardwell, V. J. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell Biol. 26, 6880–6889 (2006).
Xiao, T. et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17, 654–663 (2003).
Krogan, N. J. et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 23, 4207–4218 (2003).
Li, B., Howe, L., Anderson, S., Yates, J. R., 3rd & Workman, J. L. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278, 8897–8903 (2003).
Li, J., Moazed, D. & Gygi, S. P. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277, 49383–49388 (2002).
Schaft, D. et al. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31, 2475–2482 (2003).
Sun, X. J. et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 280, 35261–35271 (2005).
Keogh, M. C. et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 (2005).
Carrozza, M. J. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005).
Joshi, A. A. & Struhl, K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20, 971–978 (2005).
Li, H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 (2006).
Pena, P. V. et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006).
Santos-Rosa, H. et al. Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 (2002).
Pokholok, D. K. et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 (2005).
Bannister, A. J. et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280, 17732–17736 (2005).
Lee, J. W., Choi, H. S., Gyuris, J., Brent, R. & Moore, D. D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 9, 243–254 (1995).
Klose, R. J. et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 (2006).
Cloos, P. A. et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311 (2006).
Whetstine, J. R. et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 (2006).
Fodor, B. D. et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 (2006). References 77–80 show that the JHDM3/JMJD2 histone demethylase enzymes are capable of removing the tri-methyl-modification state.
Zhang, D., Yoon, H. G. & Wong, J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell Biol. 25, 6404–6414 (2005).
Vakoc, C. R. et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462 (2005).
Vakoc, C. R., Sachdeva, M. M., Wang, H. & Blobel, G. A. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell Biol. 26, 9185–9195 (2006).
Yang, Z. Q. et al. Identification of a novel gene, GASC1, within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 60, 4735–4739 (2000).
Klose, R. J. et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 22 Feb 2007 (doi: 10.1016/j.cell.2007.02.013).
Lee, N. et al. The trithorax group protein Lid is a trimethyl-H3K4 demethylase. Nature Struct. Mol. Biol. (in the press).
Liang, G., Klose, R. J., Gardener, K. E. & Zhang, Y. Yeast Jhd2 is an H3-K4 tri-methyl demethylase. Nature Struct. Mol. Biol. 18 Feb 2007 (doi: 10.1038/nsmb1204). References 85–87 show that the JARID1 family of JmjC-domain-containing proteins are H3K4 demethylases with the capacity to remove the tri-methyl-modification state.
Lu, P. J. et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274, 15633–15645 (1999).
Jensen, L. R. et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236 (2005).
Gildea, J. J., Lopez, R. & Shearn, A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156, 645–663 (2000).
Clifton, I. J. et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins. J. Inorg. Biochem. 100, 644–669 (2006).
Chen, Z. et al. Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702 (2006). Reports the first crystal structure of the enzymatic domain of an active JmjC-domain-containing histone demethylase, JHDM3A/JMJD2A.
Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002).
Muller, J. et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 (2002).
Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 (2002).
Czermin, B. et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 (2002).
Boyer, L. A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006).
Issaeva, I. et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell Biol. (2006).
Klymenko, T. & Muller, J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5, 373–377 (2004).
Papp, B. & Muller, J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 20, 2041–2054 (2006).
Poux, S., Horard, B., Sigrist, C. J. & Pirrotta, V. The Drosophila Trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development 129, 2483–2493 (2002).
Zhang, L., Schroeder, S., Fong, N. & Bentley, D. L. Altered nucleosome occupancy and histone H3K4 methylation in response to 'transcriptional stress'. EMBO J. 24, 2379–2390 (2005).
Katan-Khaykovich, Y. & Struhl, K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24, 2138–2149 (2005).
Morillon, A., Karabetsou, N., Nair, A. & Mellor, J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18, 723–734 (2005).
van Leeuwen, F., Gafken, P. R. & Gottschling, D. E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002).
Ng, H. H., Ciccone, D. N., Morshead, K. B., Oettinger, M. A. & Struhl, K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl Acad. Sci. USA 100, 1820–1825 (2003).
Schotta, G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004).
Kourmouli, N. et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117, 2491–2501 (2004).
Sims, J. K., Houston, S. I., Magazinnik, T. & Rice, J. C. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J. Biol. Chem. 281, 12760–12766 (2006).
Dunwell, J. M., Culham, A., Carter, C. E., Sosa-Aguirre, C. R. & Goodenough, P. W. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26, 740–746 (2001).
Clissold, P. M. & Ponting, C. P. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem. Sci. 26, 7–9 (2001).
Chinenov, Y. A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem. Sci. 27, 115–117 (2002).
Paraskevopoulou, C., Fairhurst, S. A., Lowe, D. J., Brick, P. & Onesti, S. The elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 59, 795–806 (2006).
Marsh, E. N., Patwardhan, A. & Huhta, M. S. S-adenosylmethionine radical enzymes. Bioorg. Chem. 32, 326–340 (2004).
Laumonnier, F. et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42, 780–786 (2005).
Ladurner, A. G. Rheostat control of gene expression by metabolites. Mol. Cell 24, 1–11 (2006).
Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000).
Landry, J. et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA 97, 5807–5811 (2000).
Tanner, K. G., Landry, J., Sternglanz, R. & Denu, J. M. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl Acad. Sci. USA 97, 14178–14182 (2000).
Lee, T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Torres-Padilla, M. E., Parfitt, D. E., Kouzarides, T. & Zernicka-Goetz, M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218 (2007).
Dodge, J. E., Kang, Y. K., Beppu, H., Lei, H. & Li, E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell Biol. 24, 2478–2486 (2004).
Tachibana, M. et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002).
O'Carroll, D. et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell Biol. 21, 4330–4336 (2001).
Luo, Y. et al. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry 45, 11727–11736 (2006).
Stone, E. M., Schaller, T. H., Bianchi, H., Person, M. D. & Fast, W. Inactivation of two diverse enzymes in the amidinotransferase superfamily by 2-chloroacetamidine: dimethylargininase and peptidylarginine deiminase. Biochemistry 44, 13744–13752 (2005).
Lee, M. G., Wynder, C., Schmidt, D. M., McCafferty, D. G. & Shiekhattar, R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13, 563–567 (2006).
Culhane, J. C. et al. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 128, 4536–4537 (2006).
Forneris, F. et al. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J. Biol. Chem. 281, 35289–35295 (2006).
Klose, R. J. et al. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol. Cell Biol. (in the press).
Seward, D. J. et al. Demethylation of trimethylated histone H3 Lys 4 in vivo by JARID1 JmjC proteins. Nature Struct. Mol. Biol. 18 Feb 2007 (doi: 10.1038/nsmb1200).
Secombe, J., Li, L., Carlos, L. & Eisenman, R. N. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 20 Feb 2007 (doi: 10.1101/gad.1523007).
Christensen, J. et al. RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell 22 Feb 2007 (doi: 10.1016/j.cell.2007.02.003).
Lee, M. G., Norman, J., Shilatifard, A. & Shiekhattar, R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a Polycomb-like protein. Cell 22 Feb 2007 (doi: 10.1016/j.cell.2007.02.004).
Iwase, S. et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 22 Feb 2007 (doi: 10.1016/j.cell.2007.02.017).
Acknowledgements
We would like to thank H. Yu, G. Zhang, R. Xu and Y. Huang for providing structural images. We also acknowledge K. Gardner for critical reading of the manuscript. We apologize to colleagues whose work we were not able to cover due to space limitations. R.J.K is funded by the Canadian Institutes of Health Research. Work in the Zhang laboratory is funded by grants from the National Institutes of Health and Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- SET domain
-
A sequence motif (named after Su(var)3–9, Enhancer of Zeste, Trithorax) that is found in several chromatin-associated proteins, including members of both the Trithorax group and Polycomb group.
- SANT domain
-
The SANT domain (named after 'switching-defective protein 3 (Swi3), adaptor 2 (Ada2), nuclear receptor co-repressor (N-CoR), transcription factor ((TF)IIIB)) is a 50-amino-acid motif that is present in nuclear receptor co-repressors and many chromatin-remodelling complexes.
- PHD domain
-
(Plant homeodomain). A zinc-binding domain found in many chromatin-associated proteins. Some PHD-domain-containing proteins have been shown to recognize methylated lysine residues in chromatin.
- Tudor domain
-
A repeated domain first identified in the Drosophila melanogaster Tudor protein, which has subsequently been identified in other proteins as a domain capable of mediating protein–nucleotide and protein–protein interactions. Recently, some Tudor domains have been shown to specifically associate with methylated lysine residues.
- X-linked mental retardation
-
A term broadly used in reference to a group of inherited mental retardations with primary genetic defects mapping to the X chromosome.
- Trithorax
-
Antagonists of Polycomb-group (PcG) proteins that maintain the active state of gene expression, whereas PcG proteins counteract this activation by repressing gene expression.
- Polycomb group
-
(PcG). A class of proteins, originally described in Drosophila melanogaster, that maintain the stable and heritable repression of several genes, including the homeotic genes.
- S-adenosylmethionine
-
(SAM). A biological compound that is involved in methyl-group transfer in living cells.
Rights and permissions
About this article
Cite this article
Klose, R., Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8, 307–318 (2007). https://doi.org/10.1038/nrm2143
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2143
This article is cited by
-
Genome-wide identification of Brassicaceae histone modification genes and their responses to abiotic stresses in allotetraploid rapeseed
BMC Plant Biology (2023)
-
Genome-wide identification and expression analysis of the JMJ-C gene family in melon (Cucumis melo L.) reveals their potential role in fruit development
BMC Genomics (2023)
-
Proline uptake promotes activation of lymphoid tissue inducer cells to maintain gut homeostasis
Nature Metabolism (2023)
-
Arabidopsis histone H3 lysine 9 methyltransferases KYP/SUVH5/6 are involved in leaf development by interacting with AS1-AS2 to repress KNAT1 and KNAT2
Communications Biology (2023)
-
Structural basis of paralog-specific KDM2A/B nucleosome recognition
Nature Chemical Biology (2023)