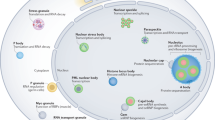
Abstract
The composition of cytoplasmic messenger ribonucleoproteins (mRNPs) is determined by their nuclear and cytoplasmic histories and reflects past functions and future fates. The protein components of selected mRNP complexes promote their assembly into microscopically visible cytoplasmic RNA granules, including stress granules, processing bodies and germ cell (or polar) granules. We propose that RNA granules can be both a cause and a consequence of altered mRNA translation, decay or editing. In this capacity, RNA granules serve as key modulators of post-transcriptional and epigenetic gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Crick, F. Central dogma of molecular biology. Nature 227, 561–563 (1970).
Sharp, P. A. The centrality of RNA. Cell 136, 577–580 (2009).
Gallois-Montbrun, S. et al. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J. Virol. 82, 5636–5642 (2008).
Gallois-Montbrun, S. et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81, 2165–2178 (2007).
Kozak, S. L., Marin, M., Rose, K. M., Bystrom, C. & Kabat, D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281, 29105–29119 (2006).
Wichroski, M., Robb, G. & Rana, T. M. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2, e41 (2006).
Goodier, J. L. & Kazazian, H. H. Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135, 23–35 (2008).
Brennecke, J. et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392 (2008).
Yin, H. & Lin, H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450, 304–308 (2007).
Nagamori, I. & Sassone-Corsi, P. The chromatoid body of male germ cells: epigenetic control and miRNA pathway. Cell Cycle 7, 3503–3508 (2008).
Rajyaguru, P. & Parker, R. CGH-1 and the control of maternal mRNAs. Trends Cell Biol. 19, 24–28 (2009).
Moore, M. J. From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518 (2005).
Sonenberg, N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 86, 178–183 (2008).
Shyu, A. B., Wilkinson, M. F. & van Hoof, A. Messenger RNA regulation: to translate or to degrade. EMBO J. 27, 471–481 (2008).
Anderson, P. & Kedersha, N. RNA granules. J. Cell Biol. 172, 803–808 (2006).
Rodriguez, A. J., Czaplinski, K., Condeelis, J. S. & Singer, R. H. Mechanisms and cellular roles of local protein synthesis in mammalian cells. Curr. Opin. Cell Biol. 20, 144–149 (2008).
Johnstone, O. & Lasko, P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35, 365–406 (2001).
Parker, R. & Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 (2007).
Anderson, P. & Kedersha, N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 (2008).
Franks, T. & Lykke-Andersen, J. The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615 (2008).
Eulalio, A., Behm-Ansmant, I. & Izaurralde, E. P bodies: at the crossroads of post-transcriptional pathways. Nature Rev. Mol. Cell Biol. 8, 9–22 (2007).
Gilks, N. et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 (2004).
Lopez de Silanes, I. et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 25, 9520–9531 (2005).
Mazan-Mamczarz, K., Lal, A., Martindale, J. L., Kawai, T. & Gorospe, M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26, 2716–2727 (2006).
Kedersha, N. L., Gupta, M., Li, W., Miller, I. & Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1441 (1999).
Kedersha, N. et al. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210 (2002).
Tourriere, H. et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 (2003).
Soncini, C., Berdo, I. & Draetta, G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20, 3869–3879 (2001).
Kraft, C., Deplazes, A., Sohrmann, M. & Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nature Cell Biol. 10, 602–610 (2008).
Ohn, T., Kedersha, N., Hickman, T., Tisdale, S. & Anderson, P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nature Cell Biol. 10, 1224–1231 (2008).
Kwon, S., Zhang, Y. & Matthias, P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381–3394 (2007).
Solomon, S. et al. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27, 2324–2342 (2007).
Richter, J. D. CPEB: a life in translation. Trends Biochem. Sci. 32, 279–285 (2007).
Wilczynska, A., Aigueperse, C., Kress, M., Dautry, F. & Weil, D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118, 981–992 (2005).
Fenger-Gron, M., Fillman, C., Norrild, B. & Lykke-Andersen, J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20, 905–915 (2005).
Wu, L., Fan, J. & Belasco, J. G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA 103, 4034–4039 (2006).
Yamashita, A. et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nature Struct. Mol. Biol. 12, 1054–1063 (2005).
Chu, C. Y. & Rana, T. M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4, e210 (2006).
Eulalio, A., Behm-Ansmant, I., Schweizer, D. & Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970–3981 (2007).
Decker, C. J., Teixeira, D. & Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 (2007).
Reijns, M. A., Alexander, R. D., Spiller, M. P. & Beggs, J. D. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463–2472 (2008).
Brengues, M., Teixeira, D. & Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489 (2005).
Hoyle, N. P., Castelli, L. M., Campbell, S. G., Holmes, L. E. & Ashe, M. P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179, 65–74 (2007).
Buchan, J. R., Muhlrad, D. & Parker, R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455 (2008).
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I. & Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 (2006).
Gallo, C. M., Munro, E., Rasoloson, D., Merritt, C. & Seydoux, G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 323, 76–87 (2008).
Jud, M. C. et al. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 318, 38–51 (2008).
Noble, S. L., Allen, B. L., Goh, L. K., Nordick, K. & Evans, T. C. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559–572 (2008).
Boag, P. R., Atalay, A., Robida, S., Reinke, V. & Blackwell, T. K. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182, 543–557 (2008).
Thomson, T., Liu, N., Arkov, A., Lehmann, R. & Lasko, P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 125, 865–873 (2008).
Lin, M. D. et al. Drosophila processing bodies in oogenesis. Dev. Biol. 322, 276–288 (2008).
Kotaja, N. et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl Acad. Sci. USA 103, 2647–2652 (2006).
Kotaja, N. & Sassone-Corsi, P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nature Rev. Mol. Cell Biol. 8, 85–90 (2007).
Kedersha, N. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 (2005).
Barbee, S. A. et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009 (2006).
Kedersha, N. et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268 (2000).
Zheng, D. et al. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182, 89–101 (2008).
Sheth, U. & Parker, R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125, 1095–1109 (2006).
Aizer, A. et al. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 19, 4154–4166 (2008).
Leatherman, J. L. & Jongens, T. A. Transcriptional silencing and translational control: key features of early germline development. Bioessays 25, 326–335 (2003).
Schisa, J. A., Pitt, J. N. & Priess, J. R. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128, 1287–1298 (2001).
Batista, P. J. et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008).
Wang, G. & Reinke, V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18, 861–867 (2008).
Nover, L., Scharf, K. D. & Neumann, D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9, 1298–1308 (1989).
Kedersha, N. & Anderson, P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969 (2002).
Stohr, N. et al. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175, 527–534 (2006).
Liu, J., Valencia-Sanchez, M. A., Hannon, G. J. & Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature Cell Biol. 7, 719–723 (2005).
Pillai, R. S. et al. Inhibition of translational initiation by let-7 microRNA in human cells. Science 309, 1573–1576 (2005).
Sen, G. L. & Blau, H. M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nature Cell Biol. 7, 633–636 (2005).
Leung, A. K. & Sharp, P. A. Function and localization of microRNAs in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 71, 29–38 (2006).
Pauley, K. M. et al. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 7, 904–910 (2006).
Goodier, J. L., Zhang, L., Vetter, M. R. & Kazazian, H. H. Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483 (2007).
Scadden, A. D. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol. Cell 28, 491–500 (2007).
Chang, W. et al. Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid–protein complex that contains mRNAs and subunits of the RNA-processing body. RNA 14, 491–502 (2008).
Ivanov, P. A., Chudinova, E. M. & Nadezhdina, E. S. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290, 227–233 (2003).
Jing, Q. et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120, 623–634 (2005).
Zeitelhofer M. et al. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J. Neurosci. 28, 7555–7562 (2008).
Acknowledgements
We thank T. K. Blackwell for critical reading of the manuscript and G. Seydoux for helpful suggestions. This work was supported by grants from the National Institutes of Health and the American College of Rheumatology.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- CAR-1
-
(Cytokinesis, apoptosis, RNA-associated 1). An Sm-like domain-containing protein that has orthologues in mammals (RAP55), Drosophila melanogaster (TRAL) and Caenorhabditis elegans (CAR-1) RNA granules.
- EDC3
-
(Enhancer of mRNA-decapping protein 3). A protein that is found in mammalian (EDC3) and yeast (Edc3) RNA granules.
- eIF3
-
(Eukaryotic translation initiation factor 3). A multisubunit complex that serves as an adaptor between eIF2, eIF4G and the small ribosomal subunit, thus facilitating initiation and stabilizing the closed loop of polysomal mRNA. eIF3 is a key component of stress granules.
- GW182
-
A large, multidomain GW repeat-containing metazoan protein that is associated with microRNAs (miRNAs) and is required for miRNA-induced gene silencing. Knockdown of GW182 inhibits the assembly of processing bodies.
- PABP1
-
(Poly(A)-binding protein 1). A protein with orthologues in mammalian (PABP1) and Caenorhabditis elegans (PAB-1) stress granules and yeast EGP bodies (Pbp1).
- PAT1
-
A translational repressor or enhancer of decapping orthologues that is found in mammalian (PAT1), yeast (Pat1) and Caenorhabditis elegans (PATR-1) RNA granules.
- PGL-1
-
(P granule abnormality 1). A protein that is found in germ cell (or polar) granules that are adjacent to nuclear pores.
- RCK
-
An RNA DEAD-box helicase that has orthologues in mammals (RCK), yeast (Dhh1), Drosophila melanogaster (ME31B) and Caenorhabditis elegans (CGH-1) RNA granules. These promote translational arrest, polysome disassembly and decapping.
- TTP
-
(Tristetraprolin). A zinc-finger-containing protein that promotes the decay of AU-rich element (ARE)-containing mRNAs at processing bodies.
Rights and permissions
About this article
Cite this article
Anderson, P., Kedersha, N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10, 430–436 (2009). https://doi.org/10.1038/nrm2694
Issue Date:
DOI: https://doi.org/10.1038/nrm2694
This article is cited by
-
ISR inhibition reverses pancreatic β-cell failure in Wolfram syndrome models
Cell Death & Differentiation (2024)
-
TOB1 and TOB2 mark distinct RNA processing granules in differentiating lens fiber cells
Journal of Molecular Histology (2024)
-
Maternal patterns of inheritance alter transcript expression in eggs
BMC Genomics (2023)
-
PDGF-BB-derived supramolecular hydrogel for promoting skin wound healing
Journal of Nanobiotechnology (2022)
-
ALBA proteins confer thermotolerance through stabilizing HSF messenger RNAs in cytoplasmic granules
Nature Plants (2022)