Key Points
-
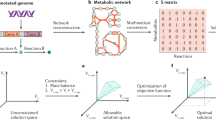
The range of allowable biochemical network functions is constrained by natural law. These constraints have been classified as first, physico-chemical constraints; second, topobiological constraints; third, environmental constraints; and fourth, self-imposed regulatory constraints. These constraints can be represented mathematically, and a growing toolbox of computational analysis methods can be used to interrogate network properties.
-
The most common set of computational tools uses linear programming to identify network states that are optimal relative to a defined objective. Flux balance analysis is an example. Optimal states have been successfully used for applications such as predicting the endpoint of adaptive evolutions of Escherichia coli.
-
A more recently developed set of computational tools are used to quantify dependencies between fluxes in the network. For example, correlated reaction sets that form functional units, or unbiased modules, of a metabolic network can be determined.
-
All allowable functional states of a biochemical network can be characterized in one of two ways: first, by enumerating network-based pathways that serve as a basis set from which all possible steady-state flux distributions can be generated or second, by using Monte-Carlo sampling of all possible functional states.
-
The development of this field has matured to the point that phenotypes of altered strains can now be predicted. One such strategy, known as pOptKnock, uses gene deletions (or additions) to align the objective of an altered cell (optimal growth) with the necessary increase in the objective of the metabolic engineer (increased secretion of a particular compound). Adaptive evolution under selective pressure for growth rate is then used to select for strains that should produce the desired product at an increased rate.
-
Future directions in this field include studying the achievable concentration states of biochemical networks, the range of in vivo kinetic constants consistent with high-throughput data and the incorporation of non-linear constraints.
Abstract
Microbial cells operate under governing constraints that limit their range of possible functions. With the availability of annotated genome sequences, it has become possible to reconstruct genome-scale biochemical reaction networks for microorganisms. The imposition of governing constraints on a reconstructed biochemical network leads to the definition of achievable cellular functions. In recent years, a substantial and growing toolbox of computational analysis methods has been developed to study the characteristics and capabilities of microorganisms using a constraint-based reconstruction and analysis (COBRA) approach. This approach provides a biochemically and genetically consistent framework for the generation of hypotheses and the testing of functions of microbial cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Covert, M. W. et al. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26, 179–186 (2001).
Edwards, J. S., Covert, M. & Palsson, B. Metabolic modelling of microbes: the flux-balance approach. Environ. Microbiol. 4, 133–140 (2002).
Reed, J. L. & Palsson, B. O. Thirteen years of building constraint-based in silico models of Escherichia coli. J. Bacteriol. 185, 2692–2699 (2003).
Goodsell, D. S. The Machinery of Life (Springer, New York, 1993).
Weisz, P. B. Diffusion and chemical transformation. Science 179, 433–440 (1973).
Elowitz, M. B., Surette, M. G., Wolf, P. E., Stock, J. B. & Leibler, S. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 181, 197–203 (1999).
Werner, A. & Heinrich, R. A kinetic model for the interaction of energy metabolism and osmotic states of human erythrocytes. Analysis of the stationary “in vivo” state and of time dependent variations under blood preservation conditions. Biomed. Biochim. Acta 44, 185–212 (1985).
Hallows, K. & Knauf, P. in Cellular and Molecular Physiology of Cell Volume Regulation. (ed. Strange, K.) 3–29 (CRC, Boca Raton, 1994).
Lew, V. L. & Bookchin, R. M. Volume, pH, and ion-content regulation in human red cells: analysis of transient behavior with an integrated model. J. Membr. Biol. 92, 57–74 (1986).
Stryer, L. Biochemistry (Freeman, New York, 1988).
Ellis, R. J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26, 597–604 (2001).
Minton, A. P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 276, 10577–10580 (2001).
Hall, D. & Minton, A. P. Macromolecular crowding: qualitative and semiquantitative successes, quantitative challenges. Biochim. Biophys. Acta 1649, 127–139 (2003).
Ellis, R. J. & Minton, A. P. Cell biology: join the crowd. Nature 425, 27–28 (2003).
Danchin, A. By way of introduction: some constraints of the cell physics that are usually forgotten, but should be taken into account for in silico genome analysis. Biochimie 78, 299–301 (1996).
Huang, J., Zhang, Q. & Schlick, T. Effect of DNA superhelicity and bound proteins on mechanistic aspects of the Hin-mediated and Fis-enhanced inversion. Biophys. J. 85, 804–817 (2003).
Neidhardt, F. C., Ingraham, J. L. & Schaechter, M. Physiology of the Bacterial Cell (Sinauer Associates, Sunderland, Massachusetts, 1990).
Danchin, A., Guerdoux-Jamet, P., Moszer, I. & Nitschke, P. Mapping the bacterial cell architecture into the chromosome. Philos. Trans. R. Soc. Lond. B 355, 179–190 (2000).
Reed, J. L., Vo, T. D., Schilling, C. H. & Palsson, B. Ø. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4, R54 (2003).
Varma, A. & Palsson, B. O. Metabolic flux balancing: basic concepts, scientific and practical use. Biotechnology 12, 994–998 (1994).
Brumen, M. & Heinrich, R. A metabolic osmotic model of human erythrocytes. Biosystems 17, 155–169 (1984).
Marhl, M., Schuster, S., Brumen, M. & Heinrich, R. Modeling the interrelations between the calcium oscillations and ER membrane potential oscillations. Biophys. Chem. 63, 221–239 (1997).
Beard, D. A., Liang, S. D. & Qian, H. Energy balance for analysis of complex metabolic networks. Biophys. J. 83, 79–86 (2002). Introduces the use of thermodynamic constraints to constraint-based analysis methods, resulting in better predictions of ranges of intracellular fluxes.
Qian, H., Beard, D. A. & Liang, S. D. Stoichiometric network theory for nonequilibrium biochemical systems. Eur. J. Biochem. 270, 415–421 (2003).
Nicholls, D. G. & Ferguson, S. J. Bioenergetics 3 (Academic, San Diego, California, 2002).
Price, N. D., Papin, J. A., Schilling, C. H. & Palsson, B. Ø. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 21, 162–169 (2003).
Covert, M. W., Famili, I. & Palsson, B. Ø. Identifying constraints that govern cell behavior: a key to converting conceptual to computational models in biology? Biotechnol. Bioeng. 84, 763–772 (2003).
Rockafellar, R. T. Convex Analysis (Princeton Univ. Press, Princeton, 1970).
Famili, I. & Palsson, B. Ø. The convex basis of the left null space of the stoichiometric matrix leads to the definition of metabolically meaningful pools. Biophys. J. 85, 16–26 (2003).
Price, N. D., Papin, J. A. & Palsson, B. Ø. Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis. Genome Res. 12, 760–769 (2002).
Papin, J. A., Price, N. D., Edwards, J. S. & Palsson, B. Ø. The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy. J. Theor. Biol. 215, 67–82 (2002).
Ibarra, R. U., Edwards, J. S. & Palsson, B. Ø. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (2002).
Edwards, J. S., Ibarra, R. U. & Palsson, B. Ø. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotechnol. 19, 125–130 (2001).
Varma, A. & Palsson, B. Ø. Predictions for oxygen supply control to enhance population stability of engineered production strains. Biotechnol. Bioeng. 43, 275–285 (1994).
Burgard, A. P., Pharkya, P. & Maranas, C. D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84, 647–657 (2003). Presents a novel method for metabolic engineering by predicting knockout strains in which the objective of the metabolic engineer and the cell are coupled.
Liao, J. C., Hou, S. Y. & Chao, Y. P. Pathway analysis, engineering and physiological considerations for redirecting central metabolism. Biotechnol. Bioeng. 52, 129–140 (1996).
Bonarius, H. P. J., Schmid, G. & Tramper, J. Flux analysis of underdetermined metabolic networks: The quest for the missing constraints. Trends Biotechnol. 15, 308–314 (1997).
Kauffman, K. J., Prakash, P. & Edwards, J. S. Advances in flux balance analysis. Curr. Opin. Biotechnol. 14, 491–496 (2003). References 37 and 38 are well-written reviews that provide an introduction to flux balance analysis, one of the most common constraint-based modelling methods.
Papoutsakis, E. T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 26, 174–187 (1984).
Majewski, R. A. & Domach, M. M. Simple constrained optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 35, 732–738 (1990).
Varma, A. & Palsson, B. Ø. Metabolic capabilities of Escherichia coli: II. Optimal growth patterns. J. Theor. Biol. 165, 503–522 (1993).
Pramanik, J. & Keasling, J. D. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 56, 398–421 (1997).
Schilling, C. H. et al. Genome-scale metabolic model of Helicobacter pylori 26695. J. Bacteriol 184, 4582–4593 (2002).
Raghunathan, A. et al. In silico metabolic model and protein expression of Haemophilus influenzae strain Rd KW20 in rich medium. OMICS 8, 25–41 (2004).
Edwards, J. S. & Palsson, B. Ø. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 274, 17410–17416 (1999).
Famili, I., Forster, J., Nielsen, J. & Palsson, B. Ø. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl Acad. Sci. USA 100, 13134–13139 (2003).
Forster, J., Famili, I., Palsson, B. Ø. & Nielsen, J. Large-scale evaluation of in silico gene knockouts in Saccharomyces cerevisiae. OMICS 7, 193–202 (2003).
Forster, J., Famili, I., Fu, P., Palsson, B. Ø. & Nielsen, J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13, 244–253 (2003).
Segre, D., Vitkup, D. & Church, G. M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl Acad. Sci. USA 99, 15112–15117 (2002). Presents a new method for predicting metabolic flux distributions of knockout strains, and shows that the predictions matched experimental data better than flux balance analysis.
Varma, A. & Palsson, B. Ø. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60, 3724–3731 (1994).
Covert, M. W. & Palsson, B. Ø. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J. Biol. Chem. 277, 28058–18064 (2002).
Raamsdonk, L. M. et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nature Biotechnol. 19, 45–50 (2001).
Thorneycroft, D., Sherson, S. M. & Smith, S. M. Using gene knockouts to investigate plant metabolism. J. Exp. Bot. 52, 1593–1601 (2001).
Bouche, N. & Bouchez, D. Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. 4, 111–117 (2001).
Lee, S., Phalakornkule, C., Domach, M. M. & Grossmann, I. E. Recursive MILP model for finding all the alternate optima in LP models for metabolic networks. Comp. Chem. Eng. 24, 711–716 (2000). First use of MILP to identify alternate equivalent optimal flux distributions in metabolic networks.
Reed, J. L. & Palsson, B. Ø. Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: assessment of correlated reaction subsets that comprise network states. Genome Res. 14, 1797–1805 (2004).
Phalakornkule, C. et al. A MILP-based flux alternative generation and NMR experimental design strategy for metabolic engineering. Metab. Eng. 3, 124–137 (2001).
Mahadevan, R. & Schilling, C. H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 (2003). Develops the method of flux variability analysis to study the effects that optimal and suboptimal solutions have on the outcome of MOMA calculations and to identify equivalent pathways in metabolic networks.
Burgard, A. P. & Maranas, C. D. Optimization-based framework for inferring and testing hypothesized metabolic objective functions. Biotechnol. Bioeng. 82, 670–677 (2003).
Varma, A., Boesch, B. W. & Palsson, B. Ø. Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates. Appl. Environ. Microbiol. 59, 2465–2473 (1993).
Edwards, J. S. & Palsson, B. Ø. Robustness analysis of the Escherichia coli metabolic network. Biotechnol. Prog. 16, 927–939 (2000).
Edwards, J. S., Ramakrishna, R. & Palsson, B. Ø. Characterizing the metabolic phenotype: a phenotype phase plane analysis. Biotechnol. Bioeng. 77, 27–36 (2002).
Kauffman, K. J., Pajerowski, J. D., Jamshidi, N., Palsson, B. Ø. & Edwards, J. S. Description and analysis of metabolic connectivity and dynamics in the human red blood cell. Biophys. J. 83, 646–662 (2002).
Burgard, A. P., Nikolaev, E. V., Schilling, C. H. & Maranas, C. D. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 14, 301–312 (2004).
Papin, J. A., Price, N. D., Wiback, S. J., Fell, D. A. & Palsson, B. Ø. Metabolic pathways in the post-genome era. Trends Biochem. Sci. 28, 250–258 (2003).
Schuster, S., Fell, D. A. & Dandekar, T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nature Biotechnol. 18, 326–332 (2000). A nice introduction to the definition and uses of elementary modes for analysing biochemical networks.
Schuster, S., Dandekar, T. & Fell, D. A. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 17, 53–60 (1999).
Schilling, C. H., Schuster, S., Palsson, B. Ø. & Heinrich, R. Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era. Biotechnol. Prog. 15, 296–303 (1999).
Papin, J. A. et al. Comparison of network-based pathway analysis methods. Trends Biotechnol. 22, 400–405 (2004).
Schuster, S. & Hilgetag, C. On elementary flux modes in biochemical reaction systems at steady state. J. Biol. Syst. 2, 165–182 (1994).
Schilling, C. H., Letscher, D. & Palsson, B. Ø. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J. Theor. Biol. 203, 229–248 (2000).
Klamt, S., Stelling, J., Ginkel, M. & Gilles, E. D. FluxAnalyzer: exploring structure, pathways, and flux distributions in metabolic networks on interactive flux maps. Bioinformatics 19, 261–269 (2003).
Pfeiffer, T., Sanchez-Valdenebro, I., Nuno, J. C., Montero, F. & Schuster, S. METATOOL: for studying metabolic networks. Bioinformatics 15, 251–257 (1999).
Klamt, S. & Stelling, J. Combinatorial complexity of pathway analysis in metabolic networks. Mol. Biol. Rep. 29, 233–236 (2002).
Price, N. D., Reed, J. L., Papin, J. A., Famili, I. & Palsson, B. Ø. Analysis of metabolic capabilities using singular value decomposition of extreme pathway matrices. Biophys. J. 84, 794–804 (2003).
Stelling, J., Klamt, S., Bettenbrock, K., Schuster, S. & Gilles, E. D. Metabolic network structure determines key aspects of functionality and regulation. Nature 420, 190–193 (2002).
Van Dien, S. J. & Lidstrom, M. E. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol. Bioeng. 78, 296–312 (2002).
Papin, J. A., Price, N. D. & Palsson B. Ø. Extreme pathway lengths and reaction participation in genome-scale metabolic networks. Genome Res. 12, 1889–1900 (2002).
Schuster, S., Klamt, S., Weckwerth, W., Moldenhauer, F. & Pfeiffer, T. Use of network analysis of metabolic systems in bioengineering. Bioprocess Biosyst. Eng. 24, 363–372 (2002).
Forster, J., Gombert, A. K. & Nielsen, J. A functional genomics approach using metabolomics and in silico pathway analysis. Biotechnol. Bioeng. 79, 703–712 (2002).
Carlson, R., Fell, D. & Srienc, F. Metabolic pathway analysis of a recombinant yeast for rational strain development. Biotechnol. Bioeng. 79, 121–134 (2002).
Price, N. D., Reed, J. L., Papin, J. A., Wiback, S. J. & Palsson, B. Ø. Network-based analysis of metabolic regulation in the human red blood cell. J. Theor. Biol. 225, 185–194 (2003).
Wiback, S. J., Mahadevan, R. & Palsson, B. Ø. Reconstructing metabolic flux vectors from extreme pathways: defining the α-spectrum. J. Theor. Biol. 224, 313–324 (2003).
Wiback, S. J., Mahadevan, R. & Palsson, B. Ø. Using metabolic flux data to further constrain the metabolic solution space and predict internal flux patterns: the Escherichia coli spectrum. Biotechnol. Bioeng. 86, 317–331 (2004).
Almaas, E., Kovacs, B., Vicsek, T., Oltvai, Z. N. & Barabasi, A. L. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427, 839–843 (2004). First paper to perform uniform random sampling of the steady-state flux space to analyse the organization of genome-scale metabolic fluxes.
Wiback, S. J., Famili, I., Greenberg, H. J. & Palsson, B. Ø. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space. J. Theor. Biol. 228, 437–447 (2004).
Fong, S. S., Marciniak, J. Y. & Palsson, B. Ø. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol. 185, 6400–6408 (2003).
Fong, S. S. & Palsson, B. Ø. Metabolic gene deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nature Genet. 36, 1056–1058 (2004).
Covert, M. W., Knight, E. M., Reed, J. L., Herrgard, M. J. & Palsson, B. Ø. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429, 92–96 (2004).
Edwards, J. S. & Palsson, B. Ø. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics 1, 1 (2000).
Edwards, J. S. & Palsson, B. Ø. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl Acad. Sci. USA 97, 5528–5533 (2000).
Duarte, N. C., Herrgard, M. J. & Palsson, B. Ø. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 14, 1298–1309 (2004).
Papp, B., Pal, C. & Hurst, L. D. Metabolic network analysis of the causes and elution of enzyme dispensability in yeast. Nature 429, 661–664 (2004). Insightful use of GEMS to demonstrate that the presence of isozymes is better explained by the need for a high flux rate through a reaction, rather than by providing redundancy for an essential function. Explains why a high degree of genes are found to be non-essential under laboratory conditions.
Burgard, A. P. & Maranas, C. D. Probing the performance limits of the Escherichia coli metabolic network subject to gene additions or deletions. Biotechnol. Bioeng. 74, 364–375 (2001).
Pharkya, P., Burgard, A. P. & Maranas, C. D. Exploring the overproduction of amino acids using the bilevel optimization framework OptKnock. Biotechnol. Bioeng. 84, 887–899 (2003).
Covert, M. & Palsson, B. Ø. Constraints-based models: regulation of gene expression reduces the steady-state solution space. J. Theor. Biol. 221, 309–325 (2003).
Covert, M. W., Schilling, C. H. & Palsson, B. Regulation of gene expression in flux balance models of metabolism. J. Theor. Biol. 213, 73–88 (2001).
Price, N. D., Famili, I., Beard, D. A. & Palsson, B. Ø. Extreme pathways and Kirchhoff's second law. Biophys. J. 83, 2879–2882 (2002).
Mahadevan, R., Edwards, J. S. & Doyle, F. J. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys. J. 83, 1331–1340 (2002).
Price, N. D., Schellenberger, J. & Palsson, B. Ø. Uniform sampling of steady state flux spaces: means to design experiments and to interpret enzymopathies. Biophsy. J. (In the press).
Segel, I. H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady–State Enzyme Systems (Wiley, New York, 1975).
Gilman, A. G. et al. Overview of the alliance for cellular signaling. Nature 420, 703–706 (2002).
Zhu, H. & Snyder, M. “Omic” approaches for unraveling signaling networks. Curr. Opin. Cell Biol. 14, 173–179 (2002).
Graves, P. R. & Haystead, T. A. A functional proteomics approach to signal transduction. Recent Prog. Horm. Res. 58, 1–24 (2003).
Li, J. et al. The molecule pages database. Nature 420, 716–717 (2002).
Sivakumaran, S., Hariharaputran, S., Mishra, J. & Bhalla, U. S. The database of quantitative cellular signaling: management and analysis of chemical kinetic models of signaling networks. Bioinformatics 19, 408–415 (2003).
Walhout, A. J. et al. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12, 1952–1958 (2002).
Papin, J. A. & Palsson, B. O. The JAK–STAT signaling network in the human B-cell: an extreme signaling pathway analysis. Biophsy. J. 87, 37–46 (2004).
Allen, T. E. & Palsson, B. Ø. Sequenced-based analysis of metabolic demands for protein synthesis in prokaryotes. J. Theor. Biol. 220, 1–18 (2003).
Lovley, D. R. Cleaning up with genomics: applying molecular biology to bioremediation. Nature Rev. Microbiol. 1, 35–44 (2003).
Edwards, J. S. & Kauffman, K. J. Biochemical engineering in the 21st century. Curr. Opin. Biotechnol. 14, 451–453 (2003).
Schilling, C. H., Edwards, J. S., Letscher, D. & Palsson, B. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol. Bioeng. 71, 286–306 (2000).
Vo, T. D., Greenberg, H. J. & Palsson, B. Ø. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J. Biol. Chem. 279, 39532–35940 (2004).
Acknowledgements
The authors would like to acknowledge funding from the United States National Institutes of Health and the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
B.Ø.P. is on the Scientific Advisory Board of Genomatica, Inc.
Related links
Glossary
- COBRA
-
(Constraint-based reconstruction and analysis). The overall philosophy and approach of applying constraints to limit the range of achievable functional (phenotypic) states of GENREs.
- CONVEX SPACE
-
A convex space is one that satisfies the following condition: given any two points in the space, the line segment in between the points is completely contained in the space. Examples of convex objects include a square, triangle or circle.
- CONCAVE SPACE
-
A space that is not convex. Examples of concave objects include a doughnut shape or a crescent.
- GENRE
-
(Genome-scale network reconstruction). Applies to a particular organism, for example, GENRE of Escherichia coli. A GENRE contains a list of all the chemical transformations that take place in the particular network. These transformations can be represented stoichiometrically. These stoichiometric representations form a matrix, the rows of which represent the compounds, the columns of which represent the chemical transformations and the entries of which are the stoichiometric coefficients.
- GEMS
-
Genome-scale models in silico of a particular organism, for example, GEMS of E. coli. The COBRA approach is used to analyse the properties of GENREs by assessing allowable functional states.
- REDUCED COST
-
A mathematical programming term; it is the smallest change in the objective function coefficient needed for a zero variable to become a non-zero variable.
- SHADOW PRICE
-
A mathematical programming term; it is the rate at which the objective value changes by increasing the supply of a particular resource (for example, a metabolite).
Rights and permissions
About this article
Cite this article
Price, N., Reed, J. & Palsson, B. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol 2, 886–897 (2004). https://doi.org/10.1038/nrmicro1023
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1023
This article is cited by
-
Genome-scale metabolic network model and phenome of solvent-tolerant Pseudomonas putida S12
BMC Genomics (2024)
-
Addressing uncertainty in genome-scale metabolic model reconstruction and analysis
Genome Biology (2021)
-
Fine tuning the glycolytic flux ratio of EP-bifido pathway for mevalonate production by enhancing glucose-6-phosphate dehydrogenase (Zwf) and CRISPRi suppressing 6-phosphofructose kinase (PfkA) in Escherichia coli
Microbial Cell Factories (2021)
-
Bayesian genome scale modelling identifies thermal determinants of yeast metabolism
Nature Communications (2021)
-
Genetic circuit characterization by inferring RNA polymerase movement and ribosome usage
Nature Communications (2020)