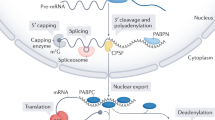
Abstract
Both end structures of eukaryotic mRNAs, namely the 5′ cap and 3′ poly(A) tail, are necessary for transcript stability, and loss of either is sufficient to stimulate decay. mRNA turnover is classically thought to be initiated by deadenylation, as has been particularly well described in Saccharomyces cerevisiae. Here we describe two additional, parallel decay pathways in the fission yeast Schizosaccharomyces pombe. First, in fission yeast mRNA decapping is frequently independent of deadenylation. Second, Cid1-dependent uridylation of polyadenylated mRNAs, such as act1, hcn1 and urg1, seems to stimulate decapping as part of a novel mRNA turnover pathway. Accordingly, urg1 mRNA is stabilized in cid1Δ cells. Uridylation and deadenylation act redundantly to stimulate decapping, and our data suggest that uridylation-dependent decapping is mediated by the Lsm1–7 complex. As human cells contain Cid1 orthologs, uridylation may form the basis of a widespread, conserved mechanism of mRNA decay.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Decker, C.J. & Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7, 1632–1643 (1993).
Muhlrad, D., Decker, C.J. & Parker, R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes Dev. 8, 855–866 (1994).
Muhlrad, D., Decker, C.J. & Parker, R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15, 2145–2156 (1995).
Anderson, J.S. & Parker, R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17, 1497–1506 (1998).
Shyu, A.B., Belasco, J.G. & Greenberg, M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5, 221–231 (1991).
Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R. & Grange, T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. USA 94, 5628–5633 (1997).
Wang, S.W., Norbury, C., Harris, A.L. & Toda, T. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 112, 927–937 (1999).
Wang, S.W., Toda, T., MacCallum, R., Harris, A.L. & Norbury, C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase δ or ε is inactivated. Mol. Cell. Biol. 20, 3234–3244 (2000).
Rissland, O.S., Mikulasova, A. & Norbury, C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 27, 3612–3624 (2007).
Kwak, J.E. & Wickens, M. A family of poly(U) polymerases. RNA 13, 860–867 (2007).
Trippe, R. et al. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12, 1494–1504 (2006).
Aravind, L. & Koonin, E.V. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27, 1609–1618 (1999).
Shen, B. & Goodman, H.M. Uridine addition after microRNA-directed cleavage. Science 306, 997 (2004).
Kaygun, H. & Marzluff, W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12, 794–800 (2005).
Mullen, T.E. & Marzluff, W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22, 50–65 (2008).
Pandey, N.B. & Marzluff, W.F. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7, 4557–4559 (1987).
Mertins, P. & Gallwitz, D. A single intronless action gene in the fission yeast Schizosaccharomyces pombe: nucleotide sequence and transcripts formed in homologous and heterologous yeast. Nucleic Acids Res. 15, 7369–7379 (1987).
Muhlrad, D. & Parker, R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6, 2100–2111 (1992).
Lackner, D.H. et al. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell 26, 145–155 (2007).
Rissland, O.S. & Norbury, C.J. The Cid1 poly(U) polymerase. Biochim. Biophys. Acta 1779, 286–294 (2008).
Sakuno, T. et al. Decapping reaction of mRNA requires Dcp1 in fission yeast: its characterization in different species from yeast to human. J. Biochem. 136, 805–812 (2004).
West, S., Gromak, N., Norbury, C.J. & Proudfoot, N.J. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell 21, 437–443 (2006).
Santiago, T.C., Purvis, I.J., Bettany, A.J. & Brown, A.J. The relationship between mRNA stability and length in Saccharomyces cerevisiae . Nucleic Acids Res. 14, 8347–8360 (1986).
Watt, S. et al. urg1: A uracil-regulatable promoter system for fission yeast with short induction and repression times. PLoS One 3, e1428 (2008).
Tucker, M., Staples, R.R., Valencia-Sanchez, M.A., Muhlrad, D. & Parker, R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae . EMBO J. 21, 1427–1436 (2002).
Tucker, M. et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell 104, 377–386 (2001).
Yamashita, A. et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12, 1054–1063 (2005).
Boeck, R. et al. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271, 432–438 (1996).
Brown, C.E., Tarun, S.Z., Jr., Boeck, R. & Sachs, A.B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae . Mol. Cell. Biol. 16, 5744–5753 (1996).
Lai, W.S., Kennington, E.A. & Blackshear, P.J. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23, 3798–3812 (2003).
Lejeune, F., Li, X. & Maquat, L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12, 675–687 (2003).
Parker, R. & Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127 (2004).
Takahashi, S., Kontani, K., Araki, Y. & Katada, T. Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1-Suc22 assembly. Nucleic Acids Res. 35, 1187–1197 (2007).
Tharun, S. et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404, 515–518 (2000).
Song, M.G. & Kiledjian, M. 3′ terminal oligo U-tract-mediated stimulation of decapping. RNA 13, 2356–2365 (2007).
Marzluff, W.F. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 17, 274–280 (2005).
Zheng, D. et al. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182, 89–101 (2008).
Sheth, U. & Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 (2003).
Brengues, M., Teixeira, D. & Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489 (2005).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol. 194, 795–823 (1991).
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast 14, 943–951 (1998).
Acknowledgements
We thank D. Bartel for his support. We also thank other members of the laboratory, A. Woollard, E. Wahle and N. Proudfoot for helpful discussions and critical reading of the manuscript, as well as T. Katada (University of Tokyo) for providing various strains. This work was supported by Cancer Research UK and the Biotechnologies and Biological Sciences Research Council. O.S.R. was supported by a scholarship from the Rhodes Trust.
Author information
Authors and Affiliations
Contributions
O.S.R. performed all the experimental work; O.S.R. and C.J.N. designed the experiments, interpreted the data and wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–4 (PDF 1623 kb)
Rights and permissions
About this article
Cite this article
Rissland, O., Norbury, C. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16, 616–623 (2009). https://doi.org/10.1038/nsmb.1601
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1601
This article is cited by
-
Membrane linked RNA glycosylation as new trend to envision epi-transcriptome epoch
Cancer Gene Therapy (2023)
-
Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression
Nature Reviews Molecular Cell Biology (2022)
-
RNA modifications: importance in immune cell biology and related diseases
Signal Transduction and Targeted Therapy (2022)
-
The TUTase URT1 connects decapping activators and prevents the accumulation of excessively deadenylated mRNAs to avoid siRNA biogenesis
Nature Communications (2021)
-
A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing
Nature Reviews Molecular Cell Biology (2020)