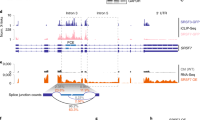
Abstract
SF2/ASF is a prototypical serine- and arginine-rich protein, with important roles in splicing and other aspects of mRNA metabolism. Splicing factor, arginine/serine-rich 1 (SFRS1), the gene encoding SF2/ASF, is a potent proto-oncogene with abnormal expression in many tumors. We found that SF2/ASF negatively autoregulates its expression to maintain homeostatic levels. We characterized six alternatively spliced SF2/ASF mRNA isoforms: the major isoform encodes full-length protein, whereas the others are either retained in the nucleus or degraded by nonsense-mediated mRNA decay. Unproductive splicing accounts for only part of the autoregulation, which occurs primarily at the translational level. The effect is specific to SF2/ASF and requires RNA recognition motif 2 (RRM2). The ultraconserved 3′ untranslated region (UTR) is necessary and sufficient for downregulation. SF2/ASF overexpression shifts the distribution of target mRNA toward monoribosomes, and translational repression is partly independent of Dicer and a 5′ cap. Thus, multiple post-transcriptional and translational mechanisms are involved in fine-tuning the expression of SF2/ASF.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wang, E.T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Cartegni, L., Chew, S.L. & Krainer, A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298 (2002).
Zhang, Z. & Krainer, A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell 16, 597–607 (2004).
Huang, Y., Gattoni, R., Stévenin, J. & Steitz, J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11, 837–843 (2003).
Lai, M.C. & Tarn, W.Y. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 279, 31745–31749 (2004).
Sanford, J.R., Gray, N.K., Beckmann, K. & Cáceres, J.F. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768 (2004).
Hanamura, A., Cáceres, J.F., Mayeda, A., Franza, B.R. Jr. & Krainer, A.R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4, 430–444 (1998).
Li, X. & Manley, J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005).
Li, X., Wang, J. & Manley, J.L. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 19, 2705–2714 (2005).
Xu, X. et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120, 59–72 (2005).
Karni, R. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 (2007).
Ghigna, C. et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890 (2005).
McGlincy, N.J. & Smith, C.W. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 33, 385–393 (2008).
Barreau, C., Paillard, L. & Osborne, H.B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150 (2005).
Valencia-Sanchez, M.A., Liu, J., Hannon, G.J. & Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524 (2006).
Morrison, M., Harris, K.S. & Roth, M.B. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94, 9782–9785 (1997).
Sureau, A., Gattoni, R., Dooghe, Y., Stévenin, J. & Soret, J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 20, 1785–1796 (2001).
Wollerton, M.C., Gooding, C., Wagner, E.J., Garcia-Blanco, M.A. & Smith, C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 (2004).
Jumaa, H. & Nielsen, P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16, 5077–5085 (1997).
Lareau, L.F., Inada, M., Green, R.E., Wengrod, J.C. & Brenner, S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446, 926–929 (2007).
Ni, J.Z. et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21, 708–718 (2007).
Bejerano, G. et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004).
Sandelin, A. et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5, 99 (2004).
Dickins, R.A. et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37, 1289–1295 (2005).
Ge, H., Zuo, P. & Manley, J.L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 66, 373–382 (1991).
Krainer, A.R., Mayeda, A., Kozak, D. & Binns, G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66, 383–394 (1991).
Isken, O. & Maquat, L.E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21, 1833–1856 (2007).
Tanguay, R.L. & Gallie, D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 16, 146–156 (1996).
Pestova, P.V., Lorsch, J.R. & Hellen, C. The mechanism of translation initiation in eukaryotes. in Translational Control in Biology and Medicine (eds. Mathews, M., Sonenberg, N. & Hershey, J.) 87–128 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2007).
Bergamini, G., Preiss, T. & Hentze, M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6, 1781–1790 (2000).
Murchison, E.P. & Hannon, G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16, 223–229 (2004).
Cummins, J.M. et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 103, 3687–3692 (2006).
Murchison, E.P., Partridge, J.F., Tam, O.H., Cheloufi, S. & Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 102, 12135–12140 (2005).
Doudna, J.A. & Sarnow, P. Translation initiation by viral internal ribosome entry sites. in Translational Control in Biology and Medicine (eds. Mathews, M., Sonenberg, N. & Hershey, J.) 129–154 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2007).
Isken, O. et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133, 314–327 (2008).
Boutz, P.L. et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21, 1636–1652 (2007).
Makeyev, E.V., Zhang, J., Carrasco, M.A. & Maniatis, T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 (2007).
Boutz, P.L., Chawla, G., Stoilov, P. & Black, D.L. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 21, 71–84 (2007).
Tacke, R., Boned, A. & Goridis, C. ASF alternative transcripts are highly conserved between mouse and man. Nucleic Acids Res. 20, 5482 (1992).
Stutz, F. & Izaurralde, E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13, 319–327 (2003).
Li, Y. et al. An Intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 443, 234–237 (2006).
Moore, M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518 (2005).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Giorgi, C. & Moore, M.J. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 18, 186–193 (2007).
Michlewski, G., Sanford, J.R. & Céceres, J.F. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E–BP1. Mol. Cell 30, 179–189 (2008).
Sanford, J.R. et al. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One 3, e3369 (2008).
Cáceres, J.F., Misteli, T., Screaton, G.R., Spector, D.L. & Krainer, A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138, 225–238 (1997).
Cazalla, D. et al. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 22, 6871–6882 (2002).
Durocher, Y., Perret, S. & Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30, E9 (2002).
Bor, Y.C. et al. Northern blot analysis of mRNA from mammalian polyribosomes. Nat. Protoc. 10.1038/nprot.2006.216 (2006).
Acknowledgements
We thank B. Vogelstein (Johns Hopkins University) for generously providing the DicerEx5/Ex5 RKO cell line, G. Hannon (Cold Spring Harbor Laboratory) for sharing the Dicer−/− and Dicer+/− ES cells, V. Racaniello (Columbia University) for the gift of HCV and CrPV IRES plasmids, C. Xue for statistical analysis, Y. Yu for advice on polysome gradients and J. Zhu for helpful discussions and communicating unpublished results. This work was supported by US National Cancer Institute grant CA13106.
Author information
Authors and Affiliations
Contributions
S.S. and A.R.K. designed the experiments and wrote the manuscript; S.S. performed the experiments and analyzed the data; Z.Z., R.S. and R.K. provided key reagents and advice.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 342 kb)
Rights and permissions
About this article
Cite this article
Sun, S., Zhang, Z., Sinha, R. et al. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol 17, 306–312 (2010). https://doi.org/10.1038/nsmb.1750
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1750
This article is cited by
-
Plant serine/arginine-rich proteins: versatile players in RNA processing
Planta (2023)
-
USP15 and USP4 facilitate lung cancer cell proliferation by regulating the alternative splicing of SRSF1
Cell Death Discovery (2022)
-
Nonsense-mediated RNA decay and its bipolar function in cancer
Molecular Cancer (2021)
-
SRSF1-dependent inhibition of C9ORF72-repeat RNA nuclear export: genome-wide mechanisms for neuroprotection in amyotrophic lateral sclerosis
Molecular Neurodegeneration (2021)
-
Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD
Nature Communications (2021)