Abstract
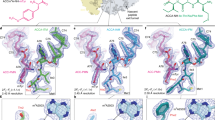
The toxin colicin E3 targets the 30S subunit of bacterial ribosomes and cleaves a phosphodiester bond in the decoding center. We present the crystal structure of the 70S ribosome in complex with the cytotoxic domain of colicin E3 (E3-rRNase). The structure reveals how the rRNase domain of colicin binds to the A site of the decoding center in the 70S ribosome and cleaves the 16S ribosomal RNA (rRNA) between A1493 and G1494. The cleavage mechanism involves the concerted action of conserved residues Glu62 and His58 of the cytotoxic domain of colicin E3. These residues activate the 16S rRNA for 2′ OH–induced hydrolysis. Conformational changes observed for E3-rRNase, 16S rRNA and helix 69 of 23S rRNA suggest that a dynamic binding platform is required for colicin E3 binding and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brodersen, D.E. et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103, 1143–1154 (2000).
Carter, A.P. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000).
Hansen, J.L. et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10, 117–128 (2002).
Auerbach, T. et al. Antibiotics targeting ribosomes: crystallographic studies. Curr. Drug Targets Infect. Disord. 2, 169–186 (2002).
Cascales, E. et al. Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 (2007).
James, R., Penfold, C.N., Moore, G.R. & Kleanthous, C. Killing of E. coli cells by E group nuclease colicins. Biochimie 84, 381–389 (2002).
Bouveret, E., Rigal, A., Lazdunski, C. & Benedetti, H. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27, 143–157 (1998).
Jakes, K.S. & Zinder, N.D. Highly purified colicin E3 contains immunity protein. Proc. Natl. Acad. Sci. USA 71, 3380–3384 (1974).
Kleanthous, C. & Walker, D. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 26, 624–631 (2001).
Walker, D., Moore, G.R., James, R. & Kleanthous, C. Thermodynamic consequences of bipartite immunity protein binding to the ribosomal ribonuclease colicin E3. Biochemistry 42, 4161–4171 (2003).
Duché, D., Frenkian, A., Prima, V. & Lloubes, R. Release of immunity protein requires functional endonuclease colicin import machinery. J. Bacteriol. 188, 8593–8600 (2006).
Bowman, C.M., Dahlberg, J.E., Ikemura, T., Konisky, J. & Nomura, M. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc. Natl. Acad. Sci. USA 68, 964–968 (1971).
Senior, B.W. & Holland, I.B. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc. Natl. Acad. Sci. USA 68, 959–963 (1971).
Soelaiman, S., Jakes, K., Wu, N., Li, C. & Shoham, M. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol. Cell 8, 1053–1062 (2001).
Carr, S., Walker, D., James, R., Kleanthous, C. & Hemmings, A.M. Inhibition of a ribosome-inactivating ribonuclease: the crystal structure of the cytotoxic domain of colicin E3 in complex with its immunity protein. Structure 8, 949–960 (2000).
Zarivach, R. et al. On the interaction of colicin E3 with the ribosome. Biochimie 84, 447–454 (2002).
Walker, D., Lancaster, L., James, R. & Kleanthous, C. Identification of the catalytic motif of the microbial ribosome inactivating cytotoxin colicin E3. Protein Sci. 13, 1603–1611 (2004).
Lancaster, L.E., Savelsbergh, A., Kleanthous, C., Wintermeyer, W. & Rodnina, M.V. Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol. Microbiol. 69, 390–401 (2008).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Voorhees, R.M., Weixlbaumer, A., Loakes, D., Kelley, A.C. & Ramakrishnan, V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 16, 528–533 (2009).
Kaufmann, Y. & Zamir, A. Protection of E. coli ribosomes against colicin E3-induced inactivation by bound aminoacyl-tRNA. FEBS Lett. 36, 277–280 (1973).
Wimberly, B.T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Neubauer, C. et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095 (2009).
Weixlbaumer, A. et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 (2008).
Korostelev, A. et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. USA 105, 19684–19689 (2008).
Sharma, D., Cukras, A.R., Rogers, E.J., Southworth, D.R. & Green, R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 374, 1065–1076 (2007).
Yusupova, G.Z., Yusupov, M.M., Cate, J.H. & Noller, H.F. The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001).
Takyar, S., Hickerson, R.P. & Noller, H.F. mRNA helicase activity of the ribosome. Cell 120, 49–58 (2005).
Yang, X., Gerczei, T., Glover, L.T. & Correll, C.C. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat. Struct. Biol. 8, 968–973 (2001).
Rupert, P.B. & Ferre-D'Amare, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410, 780–786 (2001).
Boon, T. Inactivation of ribosomes in vitro by colicin E 3. Proc. Natl. Acad. Sci. USA 68, 2421–2425 (1971).
Bowman, C.M., Sidikaro, J. & Nomura, M. Specific inactivation of ribosomes by colicin E3 in vitro and mechanism of immunity in colicinogenic cells. Nat. New Biol. 234, 133–137 (1971).
Boon, T. Inactivation of ribosomes in vitro by colicin E 3 and its mechanism of action. Proc. Natl. Acad. Sci. USA 69, 549–552 (1972).
Bowman, C.M. Inactivation of ribosomes by colicin E3 in vitro: Requirement for 50 S ribosomal subunits. FEBS Lett. 22, 73–75 (1972).
Ohno-Iwashita, Y. & Imahori, K. Colicin E3 induced cleavage of 16s rRNA of isolated 30S ribosomal subunits. J. Biochem. 82, 919–922 (1977).
Twilt, J.C., Overbeek, G.P. & van Duin, J. Translational fidelity and specificity of ribosomes cleaved by cloacin DF13. Eur. J. Biochem. 94, 477–484 (1979).
Sander, G. Colicin E3 treatment renders ribosomes more resistant to streptomycin and reduces miscoding. FEBS Lett. 97, 217–220 (1979).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Ogle, J.M., Murphy, F.V., Tarry, M.J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Ogle, J.M. & Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 74, 129–177 (2005).
Acknowledgements
We would like to thank C. Neubauer, I.S. Fernandez, Y. Gao, M. Schmeing, J. Li and M. Ortiz-Lombardia for helpful discussions and N. Kirkpatrick for technical assistance. V.R. was supported by the Medical Research Council (UK), the Wellcome Trust, the Louis-Jeantet Foundation and the Agouron Institute. C.K. acknowledges the Biotechnology and Biological Sciences Research Councils (UK) for funding.
Author information
Authors and Affiliations
Contributions
C.L.N. and K.L. carried out the crystallographic and biochemical experiments, analyzed data and interpreted results; N.A.G.M. and A.S. expressed and purified the E3-rRNase mutants; A.C.K. prepared 70S ribosomes; C.K. and V.R. supervised the research; K.L., C.L.N., C.K. and V.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 5991 kb)
Rights and permissions
About this article
Cite this article
Ng, C., Lang, K., Meenan, N. et al. Structural basis for 16S ribosomal RNA cleavage by the cytotoxic domain of colicin E3. Nat Struct Mol Biol 17, 1241–1246 (2010). https://doi.org/10.1038/nsmb.1896
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1896
This article is cited by
-
Toxin import through the antibiotic efflux channel TolC
Nature Communications (2021)
-
Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing
Biology Direct (2013)
-
A structural basis for streptomycin-induced misreading of the genetic code
Nature Communications (2013)
-
Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics
Biology Direct (2012)