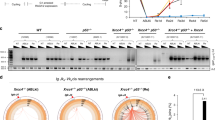
Abstract
Chromosomal translocations arise from the misjoining of DNA breaks, but the identity of the DNA repair factors and activities involved in their formation has been elusive. Here we show that depletion of CtIP, a DNA end-resection factor, results in a substantial decrease in chromosomal translocation frequency in mouse cells. Moreover, microhomology usage, a signature of the alternative nonhomologous end-joining pathway (alt-NHEJ), is significantly lower in translocation breakpoint junctions recovered from CtIP-depleted cells than in those from wild-type cells. Thus, we directly demonstrate that CtIP-mediated alt-NHEJ has a primary role in translocation formation. CtIP depletion in Ku70−/− cells reduces translocation frequency without affecting microhomology, indicating that Ku70-dependent NHEJ generates a fraction of translocations in wild-type cells. Translocations from both wild-type and Ku70−/− cells have smaller deletions on the participating chromosomes when CtIP is depleted, implicating the end-resection activity of CtIP in translocation formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lieber, M.R., Yu, K. & Raghavan, S.C. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst.) 5, 1234–1245 (2006).
Zhang, Y. & Rowley, J.D. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst.) 5, 1282–1297 (2006).
Brandt, V.L. & Roth, D.B. Recent insights into the formation of RAG-induced chromosomal translocations. Adv. Exp. Med. Biol. 650, 32–45 (2009).
Wang, J.H. et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 460, 231–236 (2009).
Robbiani, D.F. et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell 36, 631–641 (2009).
Yan, C.T. et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007).
Boboila, C. et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA 107, 3034–3039 (2010).
Nussenzweig, A. & Nussenzweig, M.C. Origin of chromosomal translocations in lymphoid cancer. Cell 141, 27–38 (2010).
Weinstock, D.M., Elliott, B. & Jasin, M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood 107, 777–780 (2006).
Stephens, P.J. et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009).
Richardson, C., Moynahan, M.E. & Jasin, M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12, 3831–3842 (1998).
Elliott, B., Richardson, C. & Jasin, M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol. Cell 17, 885–894 (2005).
Lieber, M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283, 1–5 (2008).
Weinstock, D.M., Brunet, E. & Jasin, M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat. Cell Biol. 9, 978–981 (2007).
Simsek, D. & Jasin, M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4–ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 17, 410–416 (2010).
Difilippantonio, M.J. et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404, 510–514 (2000).
Boulton, S.J. & Jackson, S.P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15, 5093–5103 (1996).
Liang, F. & Jasin, M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 271, 14405–14411 (1996).
McVey, M. & Lee, S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538 (2008).
Sartori, A.A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Clerici, M., Mantiero, D., Lucchini, G. & Longhese, M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 280, 38631–38638 (2005).
Lee, K. & Lee, S.E. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176, 2003–2014 (2007).
Bennardo, N., Cheng, A., Huang, N. & Stark, J.M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008).
Lee-Theilen, M., Matthews, A.J., Kelly, D., Zheng, S. & Chaudhuri, J. CtIP promotes microhomology-mediated alternative end-joining during class-switch recombination. Nat. Struct. Mol. Biol. advance online publication, doi:10.1038/nsmb.1942 (5 December 2010).
Rass, E. et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16, 819–824 (2009).
Xie, A., Kwok, A. & Scully, R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16, 814–818 (2009).
Mimitou, E.P. & Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008).
Zhu, Z., Chung, W.H., Shim, E.Y., Lee, S.E. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008).
Yun, M.H. & Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 (2009).
Lee, S.E. et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409 (1998).
Zhang, Y. et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14, 639–646 (2007).
Pierce, A.J., Hu, P., Han, M., Ellis, N. & Jasin, M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15, 3237–3242 (2001).
Weinstock, D.M. & Jasin, M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol. Cell. Biol. 26, 131–139 (2006).
Roth, D.B. & Wilson, J.H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 6, 4295–4304 (1986).
Blackwell, T.K. et al. Isolation of scid pre-B cells that rearrange kappa light chain genes: formation of normal signal and abnormal coding joins. EMBO J. 8, 735–742 (1989).
Brummelkamp, T.R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Yu, X. & Baer, R. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275, 18541–18549 (2000).
Acknowledgements
We thank J. Chaudhuri and colleagues (Memorial Sloan-Kettering Cancer Center) for sharing unpublished results, R. Baer (Columbia University) for reagents, and E. Brunet, D. Simsek, P. Sung and other members of the Jasin laboratory for helpful discussions. This work was supported by a Leukemia and Lymphoma Society fellowship (Y.Z.), a Dorothy Rodbell Cohen Cancer Research Program Grant and US National Institutes of Health grant R01 NIHGM54668 (M.J.).
Author information
Authors and Affiliations
Contributions
Y.Z. performed the experiments. Y.Z. and M.J. designed the research and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Table 1 (PDF 701 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Jasin, M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18, 80–84 (2011). https://doi.org/10.1038/nsmb.1940
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1940
This article is cited by
-
Ku70 affects the frequency of chromosome translocation in human lymphocytes after radiation and T-cell acute lymphoblastic leukemia
Radiation Oncology (2022)
-
ATM phosphorylates PP2A subunit A resulting in nuclear export and spatiotemporal regulation of the DNA damage response
Cellular and Molecular Life Sciences (2022)
-
Activation of homologous recombination in G1 preserves centromeric integrity
Nature (2021)
-
C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains
Nature Communications (2018)
-
Rad52 competes with Ku70/Ku86 for binding to S-region DSB ends to modulate antibody class-switch DNA recombination
Nature Communications (2017)