Abstract
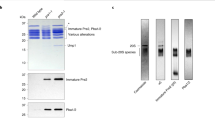
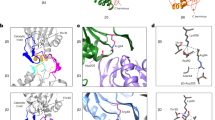
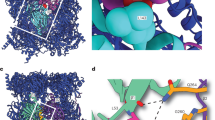
Dedicated chaperones facilitate the assembly of the eukaryotic proteasome, but how they function remains largely unknown. Here we show that a yeast 20S proteasome assembly factor, Pba1–Pba2, requires a previously overlooked C-terminal hydrophobic-tyrosine-X (HbYX) motif for function. HbYX motifs in proteasome activators open the 20S proteasome entry pore, but Pba1–Pba2 instead binds inactive proteasomal precursors. We discovered an archaeal ortholog of this factor, here named PbaA, that also binds preferentially to proteasomal precursors in a HbYX motif–dependent fashion using the same proteasomal α-ring surface pockets as are bound by activators. PbaA and the related PbaB protein can be induced to bind mature 20S proteasomes if the active sites in the central chamber are occupied by inhibitors. Our data are consistent with an allosteric mechanism in which the maturation of the proteasome active sites determines the binding of assembly chaperones, potentially shielding assembly intermediates or misassembled complexes from nonproductive associations until assembly is complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Tanaka, K. The proteasome: overview of structure and functions. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 85, 12–36 (2009).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 (1997).
Kusmierczyk, A.R. & Hochstrasser, M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol. Chem. 389, 1143–1151 (2008).
Ramos, P.C. & Dohmen, R.J. PACemakers of proteasome core particle assembly. Structure 16, 1296–1304 (2008).
Murata, S., Yashiroda, H. & Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 (2009).
Li, X., Kusmierczyk, A.R., Wong, P., Emili, A. & Hochstrasser, M. β-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 26, 2339–2349 (2007).
Hirano, Y. et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437, 1381–1385 (2005).
Le Tallec, B. et al. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 27, 660–674 (2007).
Kusmierczyk, A.R., Kunjappu, M.J., Funakoshi, M. & Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 15, 237–244 (2008).
Yashiroda, H. et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 15, 228–236 (2008).
Schmidt, M. et al. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 12, 294–303 (2005).
Marques, A.J., Glanemann, C., Ramos, P.C. & Dohmen, R.J. The C-terminal extension of the β7 subunit and activator complexes stabilize nascent 20S proteasomes and promote their maturation. J. Biol. Chem. 282, 34869–34876 (2007).
Fehlker, M., Wendler, P., Lehmann, A. & Enenkel, C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 4, 959–963 (2003).
Kaneko, T. et al. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137, 914–925 (2009).
Saeki, Y., Toh, E.A., Kudo, T., Kawamura, H. & Tanaka, K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137, 900–913 (2009).
Funakoshi, M., Tomko, R.J. Jr. Kobayashi, H. & Hochstrasser, M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137, 887–899 (2009).
Roelofs, J. et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 (2009).
Le Tallec, B., Barrault, M.B., Guerois, R., Carre, T. & Peyroche, A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell 33, 389–399 (2009).
Lowe, J. et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268, 533–539 (1995).
Maupin-Furlow, J.A. et al. Proteasomes from structure to function: perspectives from Archaea. Curr. Top. Dev. Biol. 75, 125–169 (2006).
Zwickl, P., Kleinz, J. & Baumeister, W. Critical elements in proteasome assembly. Nat. Struct. Biol. 1, 765–770 (1994).
Smith, D.M. et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007).
Rabl, J. et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 (2008).
Gillette, T.G., Kumar, B., Thompson, D., Slaughter, C.A. & DeMartino, G.N. Differential roles of the COOH termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26S proteasome. J. Biol. Chem. 283, 31813–31822 (2008).
Chen, P. & Hochstrasser, M. Biogenesis, structure and function of the yeast 20S proteasome. EMBO J. 14, 2620–2630 (1995).
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA 98, 3056–3061 (2001).
Hendrickson, E.L. et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186, 6956–6969 (2004).
Chen, P. & Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86, 961–972 (1996).
Groll, M., Brandstetter, H., Bartunik, H., Bourenkow, G. & Huber, R. Investigations on the maturation and regulation of archaebacterial proteasomes. J. Mol. Biol. 327, 75–83 (2003).
Osmulski, P.A. & Gaczynska, M. Atomic force microscopy reveals two conformations of the 20S proteasome from fission yeast. J. Biol. Chem. 275, 13171–13174 (2000).
Osmulski, P.A. & Gaczynska, M. Nanoenzymology of the 20S proteasome: proteasomal actions are controlled by the allosteric transition. Biochemistry 41, 7047–7053 (2002).
Whitby, F.G. et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408, 115–120 (2000).
Groll, M. et al. A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 (2000).
Osmulski, P.A., Hochstrasser, M. & Gaczynska, M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure 17, 1137–1147 (2009).
Kleijnen, M.F. et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 14, 1180–1188 (2007).
Forster, A., Masters, E.I., Whitby, F.G., Robinson, H. & Hill, C.P. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 (2005).
McCutchen-Maloney, S.L. et al. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 275, 18557–18565 (2000).
Zaiss, D.M., Standera, S., Holzhutter, H., Kloetzel, P. & Sijts, A.J. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 457, 333–338 (1999).
Stadtmueller, B.M. et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J. Biol. Chem. 285, 13–17 (2009).
Iyer, L.M., Burroughs, A.M. & Aravind, L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol. Direct 3, 45 (2008).
Sharon, M., Witt, S., Glasmacher, E., Baumeister, W. & Robinson, C.V. Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J. Biol. Chem. 282, 18448–18457 (2007).
Guthrie, C. & Fink, G.R. Guide to Yeast Genetics and Molecular Biology (Academic, San Diego, 1991).
Verma, R. et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 (2000).
Acknowledgements
We thank R.J. Tomko Jr. for comments on the manuscript and J. Leigh (University of Washington) for the archaeal genomic DNA. This work was supported by US National Institutes of Health grant GM083050 to M.H.
Author information
Authors and Affiliations
Contributions
A.R.K. and M.H. developed the experimental approach. A.R.K., M.J.K. and R.Y.K. carried out experiments. A.R.K. and M.H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Tables 1 and 2, Supplementary Notes and Supplementary Methods (PDF 543 kb)
Rights and permissions
About this article
Cite this article
Kusmierczyk, A., Kunjappu, M., Kim, R. et al. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat Struct Mol Biol 18, 622–629 (2011). https://doi.org/10.1038/nsmb.2027
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2027
This article is cited by
-
How to build a proteasome
Nature Structural & Molecular Biology (2021)
-
Proteasome dysfunction in alveolar type 2 epithelial cells is associated with acute respiratory distress syndrome
Scientific Reports (2019)
-
Structural insights on the dynamics of proteasome formation
Biophysical Reviews (2018)
-
Proteasome assembly from 15S precursors involves major conformational changes and recycling of the Pba1–Pba2 chaperone
Nature Communications (2015)
-
Maturation of the proteasome core particle induces an affinity switch that controls regulatory particle association
Nature Communications (2015)