Abstract
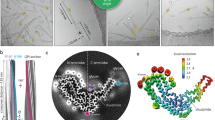
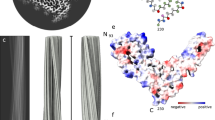
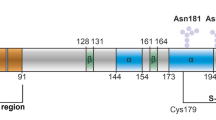
One of the mysteries in prion research is the structure of the infectious form of mammalian prion protein PrPSc. Here we used mass spectrometry analysis of hydrogen-deuterium exchange to examine brain-derived PrPSc. Our data indicate that, contrary to popular models, prion-protein conversion involves refolding of the entire region from residue ~80–90 to the C-terminus, which in PrPSc consists of β-strands and relatively short turns and/or loops, with no native α-helices present.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Prusiner, S.B. Proc. Natl. Acad. Sci. USA 95, 13363–13383 (1998).
Cobb, N.J. & Surewicz, W.K. Biochemistry 48, 2574–2585 (2009).
Castilla, J., Saa, P., Hetz, C. & Soto, C. Cell 121, 195–206 (2005).
Deleault, N.R., Harris, B.T., Rees, J.R. & Supattapone, S. Proc. Natl. Acad. Sci. USA 104, 9741–9746 (2007).
Kim, J.I. et al. J. Biol. Chem. 285, 14083–14087 (2010).
Wang, F., Wang, X., Yuan, C.G. & Ma, J. Science 327, 1132–1135 (2010).
Wüthrich, K. & Riek, R. Adv. Protein Chem. 57, 55–82 (2001).
Caughey, B.W. et al. Biochemistry 30, 7672–7680 (1991).
Safar, J., Roller, P.P., Gajdusek, D.C. & Gibbs, C.J. Jr. Protein Sci. 2, 2206–2216 (1993).
Gasset, M., Baldwin, M.A., Fletterick, R.J. & Prusiner, S.B. Proc. Natl. Acad. Sci. USA 90, 1–5 (1993).
Toyama, B.H., Kelly, M.J., Gross, J.D. & Weissman, J.S. Nature 449, 233–237 (2007).
Lu, X., Wintrode, P.L. & Surewicz, W.K. Proc. Natl. Acad. Sci. USA 104, 1510–1515 (2007).
Chesebro, B. et al. Science 308, 1435–1439 (2005).
Cobb, N.J., Sonnichsen, F.D., McHaourab, H. & Surewicz, W.K. Proc. Natl. Acad. Sci. USA 104, 18946–18951 (2007).
Govaerts, C., Wille, H., Prusiner, S.B. & Cohen, F.E. Proc. Natl. Acad. Sci. USA 101, 8342–8347 (2004).
Wille, H. et al. Proc. Natl. Acad. Sci. USA 106, 16990–16995 (2009).
DeMarco, M.L. & Daggett, V. Proc. Natl. Acad. Sci. USA 101, 2293–2298 (2004).
Spassov, S., Beekes, M. & Naumann, D. Biochim. Biophys. Acta 1760, 1138–1149 (2006).
Wickner, R.B., Shewmaker, F., Kryndushkin, D. & Edskes, H.K. Bioessays 30, 955–964 (2008).
Sawaya, M.R. et al. Nature 447, 453–457 (2007).
Acknowledgements
This study was supported by US National Institutes of Health grants NS44158, NS38604 and AG14359, and by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Contributions
V.S. conducted and analyzed H/D exchange experiments. G.J.R. and D.K.O. did all animal-associated work, from animal inoculations to dissection of brain tissue. G.S.B., D.K.O. and G.J.R. prepared PrPSc samples and carried out their biochemical characterization. B.C. did FTIR experiments. W.K.S. wrote the manuscript and coordinated the entire project. G.S.B., V.S. and B.C. discussed the results and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Methods and Supplementary Discussion (PDF 2703 kb)
Rights and permissions
About this article
Cite this article
Smirnovas, V., Baron, G., Offerdahl, D. et al. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol 18, 504–506 (2011). https://doi.org/10.1038/nsmb.2035
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2035
This article is cited by
-
Advances in mass spectrometry to unravel the structure and function of protein condensates
Nature Protocols (2023)
-
Extracellular vesicles with diagnostic and therapeutic potential for prion diseases
Cell and Tissue Research (2023)
-
Cryo-EM structure of anchorless RML prion reveals variations in shared motifs between distinct strains
Nature Communications (2022)
-
Cryo-EM structure of an amyloid fibril formed by full-length human prion protein
Nature Structural & Molecular Biology (2020)
-
Sephin1 Reduces Prion Infection in Prion-Infected Cells and Animal Model
Molecular Neurobiology (2020)