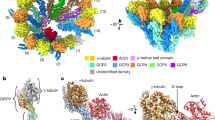
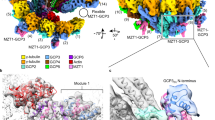
Abstract
Microtubule nucleation in all eukaryotes involves γ-tubulin small complexes (γTuSCs) that comprise two molecules of γ-tubulin bound to γ-tubulin complex proteins (GCPs) GCP2 and GCP3. In many eukaryotes, multiple γTuSCs associate with GCP4, GCP5 and GCP6 into large γ-tubulin ring complexes (γTuRCs). Recent cryo-EM studies indicate that a scaffold similar to γTuRCs is formed by lateral association of γTuSCs, with the C-terminal regions of GCP2 and GCP3 binding γ-tubulin molecules. However, the exact role of GCPs in microtubule nucleation remains unknown. Here we report the crystal structure of human GCP4 and show that its C-terminal domain binds directly to γ-tubulin. The human GCP4 structure is the prototype for all GCPs, as it can be precisely positioned within the γTuSC envelope, revealing the nature of protein-protein interactions and conformational changes regulating nucleation activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Moritz, M. & Agard, D.A. Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11, 174–181 (2001).
Kollman, J.M., Polka, J.K., Zelter, A., Davis, T.N. & Agard, D.A. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879–882 (2010).
Kollman, J.M. et al. The structure of the gamma-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell 19, 207–215 (2008).
Aldaz, H., Rice, L.M., Stearns, T. & Agard, D.A. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature 435, 523–527 (2005).
Gunawardane, R.N. et al. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J. Cell Biol. 151, 1513–1524 (2000).
Murphy, S.M. et al. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol. Biol. Cell 12, 3340–3352 (2001).
Fava, F. et al. Human 76p: A new member of the gamma-tubulin-associated protein family. J. Cell Biol. 147, 857–868 (1999).
Nakamura, M. & Hashimoto, T. A mutation in the Arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122, 2208–2217 (2009).
Knop, M., Pereira, G., Geissler, S., Grein, K. & Schiebel, E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16, 1550–1564 (1997).
Choy, R.M., Kollman, J.M., Zelter, A., Davis, T.N. & Agard, D.A. Localization and orientation of the gamma-tubulin small complex components using protein tags as labels for single particle EM. J. Struct. Biol. 168, 571–574 (2009).
Inclán, Y.F. & Nogales, E. Structural models for the self-assembly and microtubule interactions of gamma-, delta- and epsilon-tubulin. J. Cell Sci. 114, 413–422 (2001).
Masuda, H., Sevik, M. & Cande, W.Z. In vitro microtubule-nucleating activity of spindle pole bodies in fission yeast Schizosaccharomyces pombe: cell cycle-dependent activation in Xenopus cell-free extracts. J. Cell Biol. 117, 1055–1066 (1992).
Oppermann, F.S. et al. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics 8, 1751–1764 (2009).
Zhang, L., Keating, T.J., Wilde, A., Borisy, G.G. & Zheng, Y. The role of Xgrip210 in gamma-tubulin ring complex assembly and centrosome recruitment. J. Cell Biol. 151, 1525–1536 (2000).
Vérollet, C. et al. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172, 517–528 (2006).
Vogt, N., Koch, I., Schwarz, H., Schnorrer, F. & Nusslein-Volhard, C. The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963–3972 (2006).
Izumi, N., Fumoto, K., Izumi, S. & Kikuchi, A. GSK-3beta regulates proper mitotic spindle formation in cooperation with a component of the gamma-tubulin ring complex, GCP5. J. Biol. Chem. 283, 12981–12991 (2008).
Xiong, Y. & Oakley, B.R. In vivo analysis of the functions of gamma-tubulin-complex proteins. J. Cell Sci. 122, 4218–4227 (2009).
Choi, Y.K., Liu, P., Sze, S.K., Dai, C. & Qi, R.Z. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191, 1089–1095 (2010).
Vinh, D.B., Kern, J.W., Hancock, W.O., Howard, J. & Davis, T.N. Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol. Biol. Cell 13, 1144–1157 (2002).
Diederichs, K. & Karplus, P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4, 269–275 (1997).
Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
LeMaster, D.M. & Richards, F.M. 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24, 7263–7268 (1985).
Julian, M. et al. γ-Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J. Cell Sci. 105, 145–156 (1993).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Cowtan, K. & Main, P. Miscellaneous algorithms for density modification. Acta Crystallogr. D Biol. Crystallogr. 54, 487–493 (1998).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Storoni, L.C., McCoy, A.J. & Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 (2003).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Galtier, N., Gouy, M. & Gautier, C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548 (1996).
Gouet, P., Courcelle, E., Stuart, D. & Metoz, F. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1998).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Suhre, K. & Sanejouand, Y.H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 32, W610–W614 (2004).
Acknowledgements
We thank M. Wright and J.-E. Gairin (Centre de Recherche en Pharmacologie-Santé, Centre National de la Recherche Scientifique Pierre Fabre, Toulouse) for their advice and support; the staff of synchrotron beamlines PROXIMA 1 at SOLEIL Synchrotron, ID14-1, ID14-2, ID23-2 and ID29 at the European Synchrotron Radiation Facility; P. Legrand from the PROXIMA 1 beamline for his help during data collection and preliminary calculation of experimental phases; and F. Viala for her help in preparing the figures. This project was financed in part with grant 08-BLAN-0281 from the French 'Agence Nationale de la Recherche' (A.M. and L.M.) and by the Howard Hughes Medical Institute and the US National Institutes of Health grant GM31627 (J.M.K. and D.A.A.).
Author information
Authors and Affiliations
Contributions
V.G. helped optimize protein production and purification, grew the crystals and conducted diffraction data collection, analyzed the structure, prepared figures and participated in manuscript writing. M.K. optimized native and SeMet-labeled protein production and purification. L.G.-P. participated in data processing and did structure determination and refinement, analyzed the structure and helped prepare tables and figures. M.-H.R. made the constructs, produced and purified the proteins, carried out Flag pulldown experiments and prepared figures. C.C. made the constructs and carried out Flag pulldown experiments. B.R.-M. did initial purification studies. C.B. participated in protein characterization. J.M.K. analyzed structure-function relationships, carried out the fitting into the cryo-EM map of γTuSC and prepared figures. D.A.A. analyzed the data and revised the manuscript. A.M. devised the experiments, designed figures and wrote the manuscript. L.M. devised the experiments, participated in diffraction data collection, analyzed the structure, designed tables and figures and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Table 1 and Supplementary Discussion (PDF 18948 kb)
Rights and permissions
About this article
Cite this article
Guillet, V., Knibiehler, M., Gregory-Pauron, L. et al. Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat Struct Mol Biol 18, 915–919 (2011). https://doi.org/10.1038/nsmb.2083
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2083
This article is cited by
-
Effects of the protein GCP4 on gametophyte development in Arabidopsis thaliana
Protoplasma (2021)
-
Regulation of microtubule nucleation mediated by γ-tubulin complexes
Protoplasma (2017)
-
14-3-3γ Prevents Centrosome Amplification and Neoplastic Progression
Scientific Reports (2016)
-
The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle
Nature Communications (2015)
-
Microtubule nucleation at the centrosome and beyond
Nature Cell Biology (2015)