Abstract
The transcriptional co-activators YAP and TAZ are downstream targets inhibited by the Hippo tumor suppressor pathway. YAP and TAZ both possess WW domains, which are important protein–protein interaction modules that mediate interaction with proline-rich motifs, most commonly PPXY. The WW domains of YAP have complex regulatory roles as exemplified by recent reports showing that they can positively or negatively influence YAP activity in a cell and context-specific manner. In this study, we show that the WW domain of TAZ is important for it to transform both MCF10A and NIH3T3 cells and to activate transcription of ITGB2 but not CTGF, as introducing point mutations into the WW domain of TAZ (WWm) abolished its transforming and transcription-promoting ability. Using a proteomic approach, we discovered potential regulatory proteins that interact with TAZ WW domain and identified Wbp2. The interaction of Wbp2 with TAZ is dependent on the WW domain of TAZ and the PPXY-containing C-terminal region of Wbp2. Knockdown of endogenous Wbp2 suppresses, whereas overexpression of Wbp2 enhances, TAZ-driven transformation. Forced interaction of WWm with Wbp2 by direct C-terminal fusion of full-length Wbp2 or its TAZ-interacting C-terminal domain restored the transforming and transcription-promoting ability of TAZ. These results suggest that the WW domain-mediated interaction with Wbp2 promotes the transforming ability of TAZ.
Similar content being viewed by others
Introduction
The Salvador/Warts/Hippo (Hippo) tumor suppressor pathway originally defined in Drosophila melanogaster is emerging as a conserved mechanism to regulate cell proliferation and organogenesis (Harvey and Tapon, 2007; Saucedo and Edgar, 2007; Zeng and Hong, 2008). The major function of the Hippo core components is to inactivate the transcriptional co-activator Yorkie in the fly, and YAP and TAZ in mammals. The core components of the Hippo pathway consist of Hippo, Salvador, Warts and Mats in the fly, and Mst1/2, WW45, LATS1/2 and Mob, respectively, in mammals (Harvey and Tapon, 2007; Pan, 2007; Saucedo and Edgar, 2007). Hippo/Mst1/2 and Salvador/WW45 form a protein kinase complex whose activity is regulated by cell–cell contact and organ size. Upon activation, this kinase complex phosphorylates and activates another protein kinase complex formed by Warts/LATS1/2 and Mats/Mob1. The activated Warts–Mats and LATS1/2–Mob1 complex then phosphorylates Yorkie in the fly, and YAP and TAZ in mammals, respectively, at Ser residues of the HXRXXS consensus sites, leading to cytoplasmic sequestration through interaction with 14-3-3 proteins and thus inactivation of these transcriptional co-activators (Kanai et al., 2000; Zhao et al., 2007). Dissociation of Yorkie, YAP and TAZ from 14-3-3 proteins allows them to be translocated into the nucleus to activate transcriptional programs that promote cell proliferation and suppress apoptosis. Within the nucleus, Yorkie interacts with the Scalloped and Homothorax transcription factors and possibly others to mediate transcriptional programing (Bandura and Edgar, 2008; Goulev et al., 2008; Wu et al., 2008; Peng et al., 2009). Similarly, the scalloped-homologues, TEAD1–4, are transcription factors that interact with YAP and TAZ in mammals to drive the functional outcome of YAP and TAZ (Vassilev et al., 2001; Mahoney et al., 2005; Zhao et al., 2008, 2009; Chan et al., 2009). The cell cycle regulator cyclin E, antiapoptotic DIAP1 and the bantam microRNA are among the best-characterized downstream targets of Yorkie, although it is unclear whether cyclin E is a direct Yorkie target gene (Huang et al., 2005; Nolo et al., 2006; Tapon et al., 2002; Thompson and Cohen, 2006).
Mutation of core components of the Hippo pathway or overexpression of Yorkie in the fly leads to tissue overgrowth due to increased proliferation and resistance to apoptosis (Harvey and Tapon, 2007; Saucedo and Edgar, 2007). Consistent with a protissue growth role of Yorkie in the fly, YAP and TAZ are candidate oncogenes in mammals. The chromosome region containing YAP is amplified in mouse liver and mammary tumors, as well as in several human cancers (Overholtzer et al., 2006; Zender et al., 2006; Steinhardt et al., 2008). YAP protein is also overexpressed in several human cancers such as hepatocellular carcinoma (Lam-Himlin et al., 2006; Goulev et al., 2008). Overexpression of YAP in non-transformed human MCF10A mammary epithelial cells causes epithelial–mesenchymal transition (EMT), suppression of apoptosis, growth factor-independent proliferation and anchorage-independent growth in soft agar (Overholtzer et al., 2006). Transgenic overexpression of YAP in mouse liver causes tissue enlargement and eventually hepatocellular carcinoma (Dong et al., 2007). TAZ protein is overexpressed in a significant fraction of primary breast cancers and its expression levels in breast cancer cell lines correlate with cell invasiveness (Chan et al., 2008). Furthermore, expression profiling experiments have also shown that the transcript levels of TAZ and its interacting partner, TEAD4, are increased in the basal (or triple negative) breast cancer type (Richardson et al., 2006; Han et al., 2008). Functionally, like YAP, TAZ promotes EMT, cell migration, invasion, proliferation and tumorigenesis (Chan et al., 2008; Lei et al., 2008, 2009, 2009a).
TAZ contains a conserved WW domain that mediates interaction with PPXY motifs (Sudol et al., 1995; Chen et al., 1997), whereas the predominant form of YAP contains two WW domains. In three recent studies, the functional role of the Yorkie and YAP WW domains was investigated and found to confer cell and context-specific regulation (Oh and Irvine, 2009; Zhao et al., 2009; Zhang et al., 2009b). The WW domains of YAP and Yorkie were essential for their ability to transform NIH-3T3 cells and promote growth of Drosophila imaginal discs, respectively, suggesting that the WW domains interact with proteins that enhance Yorkie/YAP activity but the nature of these factors remains elusive (Zhao et al., 2009; Zhang et al., 2009b). By contrast, in MCF10A cells, mutation of the WW domains increased YAP's ability to transform MCF10A cells, suggesting that in this cell type the WW domains mediate primarily negative regulatory interactions (Zhang et al., 2009b). As many proteins other than 14-3-3 and TEAD proteins interact with YAP and/or TAZ (Wang et al., 2009), this seeming controversy likely reflects the fact that WW domains mediate a complex network of interactions with proteins that inhibit or promote Yorkie/YAP activity in cell and context-specific manners. This apparent complex regulation prompted us to examine the regulatory role of the WW domain of TAZ. Our results suggest that the WW domain of TAZ is important for its transforming potential and that Wbp2 is likely to be the key protein that interacts with the TAZ WW domain to facilitate its transforming ability.
Results
The WW domain of TAZ is critical for its ability to transform MCF10A cells
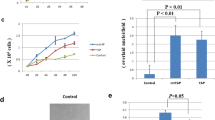
Our earlier studies have shown that TAZ is able to promote migration, invasion and is important for the transformed phenotypes of breast cancer cells (Chan et al., 2008). Anchorage-independent growth of immortalized MCF10A cells in soft agar has been used to assess the tumorigenic property of TAZ, and it was shown that interaction of TAZ with TEAD transcriptional factors is important for the transforming ability (Chan et al., 2009). To investigate the importance of the WW domain of TAZ, a WW domain mutant of TAZ (WWm) was created and examined. As shown in Figures 1a–c, MCF10A cells transduced with WWm formed significantly fewer colonies as compared with cells expressing wild-type TAZ. Consistently with earlier results, the Hippo-refractory mutant TAZ-S89A displayed enhanced transforming ability (Chan et al., 2009). These results suggest that the WW domain is important for the transforming activity of TAZ.
The WW domain of TAZ is important for its ability to confer anchorage-independent growth of MCF10A cells. (a) Soft agar growth assays were done using MCF10A cells transduced with expressing vector (control), Flag-TAZ, Flag-S89A and Flag-WWm. The colonies grown in soft agar after 1 month of culture were stained with thiazolyl blue tetrazolium bromide. (b) Lysates derived from the cells were analyzed by immunoblotting using anti-Flag antibody. (c) The colony number was quantified from three independent soft agar assays and presented as percentage relative to S89A, which is arbitrarily set as 100%. *P<0.05 versus TAZ. (d, e) The relative transcript levels of ITGB2 and CTGF in MCF10A cells stably expressing S89A, TAZ and WWm were assessed by real-time PCR. The relative expression level of ITGB2 and CTGF was presented as fold activation relative to vector control, which is set as 1. *P<0.05 versus TAZ.
The WW domain of TAZ is required for transcriptional activation of ITGB2 but not CTGF
TAZ shares several properties with YAP and there is a significant overlap among the genes that are activated by TAZ and YAP based on microarray analysis. Among the genes that are upregulated by both TAZ and YAP in a TEAD-dependent manner are ITGB2 and CTGF (Zhang et al., 2009a). It was shown that upregulation of ITGB2, but not CTGF, by YAP is dependent on the WW domains (Zhao et al., 2009). We thus assessed the transcript levels of ITGB2 and CTGF in MCF10A cells transduced with vector, TAZ, its activating mutant S89A or WWm. As shown in Figures 1d and e, TAZ induced activation of both ITGB2 and CTGF. The induction was further enhanced by the S89A mutation. However, the induction of ITGB2, but not CTGF, was compromised for WWm. The ability of WWm to activate transcription of CTGF suggests that there is no major conformational or folding change for this TAZ mutant. This result is consistent with an earlier report showing that YAP-induced upregulation of ITGB2, but not CTGF, is dependent on the YAP WW domains (Zhao et al., 2009).
Wbp2 interacts with TAZ in a WW domain-dependent manner
To identify proteins that are potential interacting partners of TAZ in a WW domain-dependent manner, we performed a large scale coimmunoprecipitation to identify the interacting proteins of TAZ (Figure 2a). Among the predominant proteins coimmunprecipitated by TAZ, S89A and WWm were various 14-3-3 protein isoforms, which were over-represented in the WWm coimmunoprecipitation as compared with S89A. Visual examination of amino-acid sequences of other coimmunoprecipitated proteins revealed several PPXY motif-containing proteins, including angiomotin (Amot), angiomotin L1 (AmotL1), Wbp2, YBX1 and Nudt21. Amot, AmotL1 and Wbp2, but not YBX1 and Nudt21, were enriched in the S89A and TAZ precipitates as compared with the WWm precipitates, suggesting that they likely interact with TAZ in a WW domain-dependent manner. As Wbp2 is known to interact with YAP (Chen and Sudol, 1995), we characterized a potential role for Wbp2 in TAZ regulation. To confirm that the interaction between Wbp2 and TAZ is dependent on the WW domain of TAZ, we transiently expressed wild-type Flag-TAZ, Flag-WWm as well as Flag-S89A into 293 cells; whole-cell lysates were immunoprecipitated with anti-Flag antibody and the immunoprecipitates were tested for the presence of Wbp2 by immunoblotting with anti-Wbp2 antibody. Figure 2b shows that both TAZ and S89A efficiently coimmunoprecipitated endogenous Wbp2, but WWm failed to interact with Wbp2. To investigate whether TAZ interacts with Wbp2 directly or through other proteins, an in vitro binding assay was performed using recombinant His-tagged Wbp2 (His-Wbp2) and glutathione S-transferase (GST)-tagged TAZ (GST-TAZ). When incubated together, His-Wbp2 can be recovered with GST-TAZ, but not GST, by glutathione beads (Figure 2c), suggesting that Wbp2 interacts with TAZ directly. These results suggest that Wbp2 interacts with TAZ directly in a WW domain-dependent manner.
Direct interaction of Wbp2 with TAZ depends on the WW domain of TAZ. (a) Several PPXY motif-containing proteins were identified by large-scale coimmunoprecipitation. A total of 293 cells expressing 3XFlag-6XHis-TAZ, -S89A and -WWm (together with cells transfected with vector) were lysed and the lysates were processed for large-scale coimmunoprecipitation. The precipitates were resolved by SDS–PAGE and the gel pieces were excised out for mass spectrometry (MS) analysis. The bands containing Amot, AmotL1, TAZ, Wbp2 and their MS coverage of the polypeptide were indicated (left). The Wbp2 peptide sequencescovered are shown (right). (b) WW domain of TAZ is required for interaction with Wbp2. Whole-cell lysates from 293 cells transiently expressing vector, Flag-TAZ, Flag-S89A and Flag-WWm were immunoprecipitated with anti-Flag, and the immunoprecipitates (together with starting lysate as input) were analyzed by immunoblot to detect coimmunoprecipitated endogenous Wbp2 (top). The blots were stripped and re-probed with anti-Flag-HRP to detect immunoprecipitated Flag-TAZ and its mutants (bottom). Mutation of WW domain of TAZ disrupted interaction with endogenous Wbp2. (c) Direct interaction of TAZ with Wbp2. GST sepharose containing equal amount of GST-TAZ or GST proteins were incubated with purified His-tagged Wbp2 protein and washed. Proteins retained on the beads were analyzed by immunoblot to detect Wbp2. His-Wbp2 is bound to GST-TAZ but not GST. (d) The purified His-Wbp2, GST and GST-TAZ proteins resolved by SDS–PAGE gel.
The interaction of Wbp2 with TAZ is mediated by its PPXY-containing C-terminal region
Alignment of amino-acid sequences of Wbp2 proteins from different species shows that there are three PPXY motifs, present in the C-terminal portion of Wbp2, and that the second and third motifs are conserved among various species such as human, zebrafish, frog, worm and fly, whereas the first motif is only conserved in some species (Supplementary Figure S1, Figure 3a). PPXY motifs have been shown to be important for binding to WW domains (Chen and Sudol, 1995; Sudol et al., 1995; Einbond and Sudol, 1996; Macias et al., 1996; Chen et al., 1997; Harvey et al., 1999). To investigate whether the PPXY motif-containing C-terminal portion of Wbp2 is important for interaction with TAZ, two truncation mutants were created, one containing the N-terminal region (amino acids 1–151) and the other containing the C-terminal region (amino acids 137–261) (Figure 3a). As the C-terminal fragment is very unstable and thus undetectable, the full-length Wbp2, N-terminal and C-terminal regions were expressed as a fusion protein with green fluorescent protein (GFP). When coexpressed with Flag-TAZ, the full-length and the C-terminal region of Wbp2 interacted with TAZ, but the N-terminal region failed to interact (Figure 3b), suggesting that the PPXY motif-containing C-terminal region of Wbp2 interacts with the WW domain of TAZ. Owing to the fact that GFP-WBP2-N-ter is expressed at much lower levels, its lack of interaction is only indicative and thus should not be interpreted alone. However, once the PPXY motifs at the C terminal of Wbp2 are mutated, the interaction between these PPXY mutants and TAZ is dramatically reduced (Figure 3c). This suggests that the C-terminal region of Wbp2 is most important for interaction with TAZ, consistent with the lack of interaction of TAZ with the GFP-WBP2-N-ter.
Interaction of Wbp2 with TAZ depends on the PPXY motifs-containing C-terminal region of Wbp2. (a) The schematic diagram shows the N-terminal, C-terminal regions of Wbp2 and the positions of three PPXY motifs. (b) The C-terminal region of Wbp2 is important for interaction with TAZ. Whole-cell lysates from 293 cells transiently expressing Flag-TAZ together with GFP-Wbp2, GFP-Wbp2-N-terminal or GFP-Wbp2-C-terminal regions were immunoprecipitated with anti-Flag, and the immunoprecipitates were analyzed by immunoblot to detect coimmunoprecipitated GFP-Wbp2 and its mutants. The blots were stripped and re-probed with anti-Flag-HRP to detect immunoprecipitated Flag-TAZ. (c) PPXY motifs (aa197-200) of Wbp2 are important for interaction with TAZ. In all, 293 cells expressing Flag-TAZ and various mutants of HA-Wbp2 were processed for immunoprecipitation. The immunoprecipitates were analyzed by immunoblot to detect coimmunoprecipitated Wbp2 and its PPXY mutants. Mutation of the second PPXY motif significantly reduced the ability of Wbp2 to interact with TAZ. Lane 1 is negative control for immunoprecipitation with 293 cells expressing vector and wild-type Wbp2 only.
The PPXY motifs of Wbp2 are important for interaction with the WW-domain of TAZ
As there are three PPXY motifs in the C-terminal portion of human Wbp2, we next investigated their relative importance in mediating interaction with TAZ. The three motifs were mutated singly, doubly in various combinations (1+2, 1+3 or 2+3), or triply (1+2+3) to generate different PPXY mutants: PPXY1, PPXY2, PPXY3, PPXY1+2, PPXY1+3, PPXY2+3 and PPXY1+2+3. These PPXY mutants, along with Flag-TAZ, were transiently expressed in 293 cells and whole-cell lysates were immunoprecipitated with anti-Flag antibody followed by immunoblotting analysis with anti-hemagglutinin (HA) antibody to detect the co-recovery of these various HA-tagged mutant proteins. Figure 3c shows that PPXY2 (lane 4), PPXY1+2 (lane 6) and PPXY1+2+3 (lane 9) are severely impaired in their property of being coimmunoprecipitated with TAZ, whereas PPXY1 (lane 3), PPXY3 (lane 5) and PPXY1+3 (lane 7) seem to have a minor effect on the interaction with TAZ. These results suggest that the second PPXY motif of Wbp2 is most important for interaction with TAZ, because its single mutation reduced the interaction to a comparable level as the mutation of all three motifs.
Knockdown of Wbp2 suppresses, whereas overexpression of Wbp2 enhances, TAZ-driven transformation
After establishing the interaction of TAZ with Wbp2, we then examined whether endogenous Wbp2 is required for TAZ-mediated transformation. We suppressed the expression of endogenous Wbp2 using a short hairpin (sh)RNA knockdown approach. As the Hippo-refractory S89A has enhanced ability to transform MCF10A cells, we used it to test the role of endogenous Wbp2 with S89A-mediated cell growth in soft agar. Soft agar growth assay was performed using MCF10A-S89A cells transduced with retroviral vector expressing shRNAs (knockdown (KD) 1 and KD2) targeting Wbp2. Both shRNAs noticeably reduced the expression of Wbp2 protein as well as its transcript in MCF10A cells, as verified by immunoblotting and quantitative PCR (Figure 4a), and reduced the colony numbers of S89A-expressing cells in soft agar (Figures 4b and c). Although Wbp2 itself did not have the ability to induce soft-agar growth in MCF10A cells, overexpression of Wbp2 enhanced TAZ-mediated transformation of MCF10A cells (Figures 4d and e). The observations that knockdown of endogenous Wbp2 caused the reduction of soft agar colony number induced by S89A and overexpression of Wbp2 enhanced TAZ-mediated transformation strongly suggest that endogenous Wbp2 is important for TAZ and its Hippo-refractory S89A mutant to transform MCF10A cells.
Suppression of Wbp2 expression inhibits, whereas overexpression of Wbp2 enhances TAZ-driven transformation of MCF10A cells. (a) The relative protein and transcript levels of Wbp2 were assessed by immunoblot and real-time PCR, respectively. The average level of Wbp2 transcript in vector-transduced cells is arbitrarily set at 1. *P<0.05 versus control. (b) S89A-driven anchorage-independent growth is suppressed by shRNA-mediated knockdown of Wbp2. S89A-expressing MCF10A cells (S89A) transduced with vector (control) or Wbp2 shRNAs (Wbp2-KD-1 or Wbp2-KD-2) targeting Wbp2 were grown in soft agar for a month and colonies were stained. (c) Quantification of soft agar colonies of S89A cells expressing Wbp2 shRNAs, the average from three independent experiments was presented. The average colony number from S89A cells expressing no shRNA targeting Wbp2 is arbitrarily set at 100%. *P<0.05 versus control. (d) Overexpression of Wbp2 enhances TAZ-driven transformation. MCF10A-vector and MCF10A-TAZ cells cotransduced with Wbp2 were used to perform soft agar assays. (e) Quantification of soft agar colonies from (d) and presented as percentage relative to TAZ+vector, which is arbitrarily set as 100%. *P<0.05 versus TAZ+control.
Forced interaction of WWm with Wbp2 by direct fusion of Wbp2 to the TAZ C-terminus restored/enhanced the transforming ability
To confirm that the reduction of transforming activity of WWm is indeed due to the loss of interaction with Wbp2, we generated a WWm–Wbp2 fusion protein by directly joining the Wbp2 to the C-terminus of WWm. As shown in Figures 5a–d, the WWm–Wbp2 fusion protein not only has restored transforming ability but also displayed a significantly enhanced ability to transform NIH3T3 cells. In fact the WWm–Wbp2 was as oncogenic as the Hippo refractory S89A mutant. The potent transforming ability exhibited by WWm–Wbp2 suggests that a reduction in the transforming ability of WWm is likely caused by its lost interaction with Wbp2, rather than by misfolding or conformation changes. Furthermore, fusion of the PPXY motif-containing C-terminal, but not the N-terminal portion of Wbp2, with WWm restored and even enhanced the transforming ability of TAZ (Figures 5a–c). Although PPXY motif-mediated interaction with TAZ is important for Wbp2 to potentiate the transforming ability of TAZ, some other regions in the C-terminal domain of Wbp2 may also be involved in the transforming activity of TAZ. In addition, the ability to induce the transcription of ITGB2 was also significantly restored for WWm when fused to Wbp2 or its C-terminal region (Figure 5d). Taken together, these results suggest that the interaction of TAZ with endogenous Wbp2 is important for TAZ to drive transcription and transform cells.
Direct fusion of full-length Wbp2 and its C-terminal, but not N-terminal region, to WWm restore the transforming and gene-induction abilities of TAZ. (a) NIH-3T3 cells stably expressing the indicated constructs grown on soft agar are shown. (b) Lysates derived from cells expressing the indicated constructs were analyzed by immunoblotting using anti-Flag antibody (left) and anti-HA antibody (right). (c) Quantification of colony numbers for all the indicated constructs in comparison with vector-transduced cells. The average number derived from S89A-expressing cells is arbitrarily set at 100%. P<0.05 versus TAZ. (d) The relative expression levels of ITGB2 in MCF10A cells stably expressing the indicated constructs were assessed by real-time PCR. *P<0.05 versus TAZ.
Discussion
In this study, we showed that the WW domain of TAZ is important for its oncogenic capability, as well as for its ability to activate transcription of genes such as ITGB2 that are known to be upregulated by TAZ. Mutation of the WW domain of TAZ reduced its transforming ability as assessed by anchorage-independent cell growth in soft agar. Our results are consistent with the finding that the YAP WW domains are required for YAP to transform NIH3T3 cells and for YAP's ability to induce ITGB2 (Zhao et al., 2009; Zhang et al., 2009b). We observed no clear role for the TAZ WW domain in mediating interactions with negative regulatory proteins, although these could have been masked if the observed positive regulatory interactions were epistatic with respect to their influence on TAZ activity. Increasing evidence shows that the WW domain of Yorkie, YAP and TAZ interacts with multiple different proteins containing PPXY motifs. Evidence from flies and from mammalian tissue culture studies suggest that the WW domains of Yorkie and YAP interact with both positive and negative regulatory proteins, and therefore it is conceivable that some WW domain-interacting proteins may also execute inhibitory effects on the transforming ability of TAZ and YAP. As such, the regulatory role of the Yorkie, YAP and TAZ WW domains may depend on the cellular context and experimental conditions that may alter the relative abundance of PPXY-containing proteins that interact with Yorkie, YAP and TAZ.
Among the proteins that were pulled down by TAZ and S89A, but not by WWm, were PPXY motif-containing Wbp2, Amot and AmotL1, implying that these proteins interact with the WW domain of TAZ. As WWm has reduced transforming ability, we explored the possibility that TAZ may engage Wbp2 via its WW domain for transforming ability and gene regulation. Consistent with this hypothesis, we provided evidence showing that Wbp2 interacts directly with TAZ and this interaction is likely mediated by the WW domain of TAZ and the second PPXY motif of Wbp2. The functional consequence of this interaction was then established by our observations that the potent transforming ability of S89A is suppressed by knockdown of endogenous Wbp2 and the moderate transforming ability of wild-type TAZ is noticeably enhanced by exogenous Wbp2, supporting a role of Wbp2 in positively regulating the transforming ability of TAZ. Further support for a role of Wbp2 came from the results that the defect of transforming ability of WWm can be rescued by fusing Wbp2 or its C-terminal region to the C-terminus of WWm. In addition, these fusion proteins displayed enhanced transforming ability, suggesting that the physical interaction of Wbp2 with TAZ may significantly empower the transforming potential of TAZ. Furthermore, mutation of the WW domain of the Hippo refractory S89A also reduced its transforming activity, whereas fusion of full-length Wbp2 to the S89A-WWm restored the activity (Supplementary Figure S2). This suggests that Wbp2 interaction with the WW domain is also involved in the much enhanced transforming activity of S89A. The enhanced transforming ability could possibly be explained by the fact that TAZ and Wbp2 would now be constitutively associated and, therefore, Wbp2 would not be subject to competition with other PPXY motif-containing proteins (especially negatively regulatory proteins) for the TAZ WW domain.
As Wbp2 is also a YAP interacting factor (Chen and Sudol, 1995), we have examined whether it can enhance the transforming ability of YAP. We first created mutant YAP having the first (YAP-WW1m), the second (YAP-WW2m) or both (YAP-WW(1+2)m) WW domains mutated and examined their transforming ability (Supplementary Figure S3). In our study, mutation of WW1, WW2 or both WW domains enhanced the transforming ability of YAP in both NIH3T3 and MCF10A cells as compared with wild-type YAP. These results are different from those reported by Guan's group, where mutations of WW domains of YAP reduced NIH3T3 cell growth in soft agar (Zhao et al., 2009), and those by Harvey's group, where YAP WW domain mutants reduce NIH3T3 cell growth but enhance MCF10A cell growth in soft agar (Zhang et al., 2009b). Although this issue remains to be further investigated, one possible explanation is that the WW domains of YAP are primarily interacting with negative factors under our experimental conditions in both NIH3T3 and MCF10A cells. Importantly, when Wbp2 was fused directly to the C terminus of these YAP mutants to create YAP-WW1m-Wbp2, YAP-WW2m-Wbp2 and YAP-WW(1+2)m-Wbp2, these fusions induced more NIH3T3 and MCF10A cell growth in soft agar as compared with their respective WW domain mutants (Supplementary Figure S3), suggesting that Wbp2 potentiates YAP transforming activity. These results suggest that physical interaction of Wbp2 with YAP also has a positive effect on its transforming ability.
YAP and TAZ are able to induce EMT and increase cell proliferation (Overholtzer et al., 2006; Lei et al., 2008; Zhang et al., 2009a, 2009b). Moreover, WW domains of YAP are essential for the increased cell proliferation, but dispensable for EMT (Zhao et al., 2009). We have also examined the relevance of the WW domain of TAZ and its interaction with Wbp2 in EMT and cell proliferation. The expression of EMT markers was examined to investigate the role of TAZ WW domain and its interaction with Wbp2 (Supplementary Figure S4A). In our study, the alterations of EMT markers are not as clear-cut as reported, although the general trend holds true. The expression of epithelial marker E-cadherin was suppressed mostly in MCF10A cells expressing S89A. E-cadherin downregulation was noticeable but only moderate in cells expressing TAZ, WWm, WWm-Wbp2 and WWm-Wbp2-C-ter, whereas cells expressing WWm-Wbp2-N-ter had similar levels of E-cadherin as vector-transduced cells. Consistently, vimentin is mostly upregulated in cells expressing WWm-Wbp2-C-ter and S89A. Significant vimentin upregulation was also observed upon overexpression of TAZ, WWm and WWm-Wbp2, but not WWm-Wbp2-N-ter. Furthermore, S89A and WWm-Wbp2-C-ter are relatively more potent in inducing the expression of fibronectin and, to a lesser extent, N-cadherin. The ability of WWm to induce the expression of some EMT markers is similar to TAZ and WWm-Wbp2. These results indicate that WW domain is not very crucial for EMT, although S89A and Wwm-Wbp2-C-ter have enhanced ability to induce EMT. Our results are consistent with the earlier observation that the WW domain of YAP is not very crucial for EMT (Zhao et al., 2009). Furthermore, cell proliferation assays show that the WW domain of TAZ (Supplementary Figure S4B) and cellular Wbp2 (Supplementary Figure S4C) are both positively involved in cell proliferation.
Consistent with the notion that the role of the WW domains of TAZ and YAP depends on the relative abundance and interacting affinity of various PPXY motif-containing proteins, our preliminary analysis indicates that Amot also interacts with the WW domain of TAZ and YAP and that this interaction seems to suppress the transforming ability of TAZ and YAP. As such, if the positive PPXY motif-containing proteins such as Wbp2 are more dominant in the cell under a certain condition, then the WW domain is important for the transforming ability. On the other hand, if the negative PPXY motif-containing proteins such as Amot are dominant, then the WW domain is involved in suppressing the transforming ability of TAZ and YAP. Future studies will provide better understanding about this hypothesis. The mechanism by which Wbp2 regulates TAZ-mediated transformation and gene expression is currently unknown. HA-Wbp2 is distributed both in the cytoplasm and in the nucleus (Supplementary Figure S5A) as assessed by immunofluorescence microscopy. When Flag-TAZ was expressed alone or together with Wbp2, TAZ is preferentially distributed in the nucleus and coexpressed Wbp2 does not have a significant effect on the localization of TAZ (Supplementary Figure S5B). Furthermore, overexpression of Wbp2 does not affect the phosphorylation or abundance of TAZ (Supplementary Figure S5C) and increase in the amount of Wbp2 does not affect the interaction of TAZ with TEAD4 (Supplementary Figure S6). Therefore, the mechanism underlying Wbp2 action needs to be further defined. A previous study has suggested that it interacts with E6-AP to promote transcriptional activation of estrogen and progesterone receptors (Dhananjayan et al., 2006). The transcriptional activation of progesterone receptor by Wbp2 may be mediated by the recruitment of YAP. It was shown that the third Wbp2 PPXY motif is most important. One possibility is that via Wbp2, other proteins such as E6-AP may be recruited to TAZ-containing complexes in such a way that transcription of selected genes such as ITGB2 is positively regulated. How the interaction of Wbp2 with TAZ is regulated is another interesting issue because it was reported that Wbp2 is tyrosine-phosphorylated in response to the stimulation by epidermal growth factor in MCF10A cells (Chen et al., 2007). It will be interesting to examine whether Tyr-phosphorylation of Wbp2 can regulate its interaction with TAZ and transcriptional outcome. Future studies addressing these issues will provide additional insights into the molecular mechanism by which TAZ and Wbp2 coordinately control transcription and cell transformation.
Materials and methods
Cell lines and materials
MCF10A and NIH-3T3 cells were purchased from the American Type Culture Collection. MCF10A cells were cultured in Dulbecco's modied Eagle's medium supplemented with 5% horse serum, 20 ng/ml of epidermal growth factor, 0.5 μg/ml of hydrocortisone, 100 ng/ml of cholera toxin and 10 μg/ml of insulin and pen/strep. NIH-3T3 cells were cultured in Dulbecco's modied Eagle's medium supplemented with 10% fetal bovine serum. Protease inhibitors cocktail was purchased from Roche Diagnostics Corporation (Roche Applied Science, Indianapolis, IN, USA). Ezview Red anti-Flag M2 affinity gel and 3XFLAG peptide were from Sigma-Aldrich (St Louis, MO, USA). Talon cobalt resin was from Clontech Laboratories, Inc. (Mountain View, CA, USA).
Antibodies
Wbp2 antibody was purchased from Abnova (Taiwan) Corporation (Taipei City, Taiwan), which recognizes endogenous Wbp2 as well as overexpressed HA-Wbp2 in MCF10A (Supplementary Figure S7). The endogenous Wbp2 exhibited an apparent size of ∼42 kDa in our system (left lane), although the predicted size of Wbp2 is about 28 kDa. The much enhanced size of the HA-Wbp2 (right lane) in the same experiment is supportive of the apparently oversized Wbp2 in the SDS–PAGE. EZview Red anti-Flag M2 affinity gel and anti-Flag M2-peroxidase (anti-Flag-horseradish peroxidase) mouse antibody were from Sigma. Anti-GFP was from Upstates Biotechnology (Millipore Corporate Headquarters, Billerica, MA, USA). Anti-HA-peroxidase (anti-HA-horseradish peroxidase) antibody was from Roche.
Plasmids
Flag-TAZ and S89A were previously described (Chan et al., 2008, 2009). Double amino-acid mutations were introduced into W152 and P155 of TAZ to create WWm (W152A, P155A) by PCR. Wbp2 complementary DNAs were obtained from MegaMan Human Trinscriptome Library (Stratagene (Agilent Technologies, Inc.), Santa Clara, CA, USA) using PCR, tagged with HA epitope at the N-terminus and cloned into pBABEpuro retroviral vector. PCR-amplified complementary DNA fragments coding for full length, the N terminal and the C terminal of Wbp2 were subcloned into pEGFP (Clontech) to make GFP-Wbp2, GFP-Wbp2-N-ter and GFP-Wbp2-C-ter. GST fusion proteins were prepared as described previously (Chan et al., 2008). WWm-Wbp2, WWm-Wbp2-N-ter and WWm-Wbp2-C-ter were constructed by fusing the full-length WWm coding region to the second codon of Wbp2, Wbp2 (amino acids 2–151) and Wbp2 (amino acids 137–261), respectively. TAZ was subcloned into pGEX-4T vector (Pharmacia, GE Healthcare Bio-Sciences AB, Uppsala, Sweden) to make GST-TAZ and Wbp2 was subcloned into pET-32 (modified) from Naiyang Fu, Singapore, to make His-Wbp2. Alanines were introduced to the Y residue of the PPXY motifs of the Wbp2-PPXY mutants by PCR. 3XFlag-6XHis-TAZ, -S89A and -WWm were cloned into pCIneo (Promega Corporation, Madison, WI, USA) vector. The sequence of the 3XFlag-6XHis is 5′-ATGGACTACAAAGACGATGACGACAAGGACTACAAAGACGATGACGACAAGGACTACAAAGACGATGACGACAAGCATCATCACCATCACCAT-3′.
Retrovirus generation and infection
The amphotropic and ecotropic Phoenix packaging cells (kindly provided by Nolan Lab at Stanford University) were transfected with the indicated retroviral vectors using Lipofectamine according to the manufacturer's instruction (Invitrogen, Carlsbad, CA, USA). After 48 h, the retroviral supernatants were collected, filtered (0.45 μm; Millipore Corporate Headquarters, Billerica, MA, USA) and added onto the target cells in the presence of 5 μg/ml of polybrene (Sigma-Aldrich) for 6–8 h. Infection was done twice. After infection, the cells were selected with puromycin (1 μg/ml) for a week before being analyzed for protein expression by immunoblotting, followed by soft agar assays. If two constructs were expressed in the cells, the transductions were done sequentially and the cells were selected with two different antibiotics, hygromycin and puromycin.
Anchorage-independent growth in soft agar
In all, 1.5 ml of 0.5% agar (electrograde ultra pure; Invitrogen) supplemented with culture media was plated in six-well plates as the bottom agar. Thirty thousand cells were mixed with 1.5 ml of 0.35% agar-supplemented media and plated on the solidified bottom agar. The media (of volume 1 ml) was added on top of the solidified agar layers and the colonies were allowed to grow in an incubator at 37 °C, 5% CO2 for a month for MCF10A cells and 2 weeks for NIH-3T3 cells. The colonies were stained with thiazolyl blue tetrazolium bromide and scanned. Colonies were quantified using Image J program developed at National Institutes of Health (NIH), USA.
RNA isolation and real-time PCR
Total RNA was isolated from culture cells using RNeasy Mini Kit form QIAGEN Sciences (Germantown, MD, USA). Complementary DNA was synthesized by reverse transcription using High Capacity complementary DNA Reverse Transcription Kit (Applied Biosystems) and subjected to real-time PCR using specific probes and reagents from Applied Biosystems. Relative abundance of mRNA was calculated using a program provided by Applied Biosystems (Life Technologies Corporation, Carlsbad, CA, USA).
shRNA-mediated knockdown of Wbp2
The sequences of the shRNAs against human Wbp2 were KD-1, 5′-AACGTGCCAGAAGCCTTCAAA-3′, and KD-2, 5′-GCGGAGTGATCGTCAATAA-3′. The shRNAs were cloned into retroviral vector pSuper.retro.puro vector (Oligoengine, Seattle, WA, USA). The relative amount of protein and transcripts for Wbp2 was assessed by immunoblotting as well as real-time PCR using reagents from Applied Biosystems.
Immunoprecipitation
Human embryonic kidney cells 293 (in a 10-cm plate) transiently transfected with the indicated plasmids were washed once with ice-cold phosphate buffered saline and subsequently lysed in 1 ml of ice-cold lysis buffer (150 mM NaCl, 50 mM Tris–HCl, pH 7.4, 0.1% NP40 and protease inhibitor cocktail (Roche)). After clearance by a spin (10 000 g, 15 min), 1 mg of whole cell lysates was incubated overnight with 70 μl of Ezview Red anti-Flag M2 affinity gel, washed twice with 1 ml of lysis buffer with 500mM NaCl and 0.5% NP40 followed by three times with lysis buffer, and finally resolved on SDS–polyacrylamide gels and blotted onto nitrocellulose membrane. The filters were blocked with 5% skimmed milk in phosphate buffered saline and probed with anti-HA-horseradish peroxidase (50 ng/ml), anti-Wbp2 (1:500) or anti-GFP (1:1000) antibodies. Signals were visualized using Supersignal (Pierce Biotechnology (Thermo Fisher Scientific), Rockford, IL, USA). The blots were stripped and re-probed with anti-Flag-horseradish peroxidase (1:1000) to detect immunoprecipitated Flag-TAZ.
Identification of interacting proteins of TAZ
293 cells transiently expressing the Flag(3X) and His(6X) tagged TAZ, S89A and WWm were used for large-scale immunoprecipitation. In all, 50 mg of whole cell lysates in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% NP40 and protease inhibitors cocktail) was incubated overnight with 1 ml of EZview Red anti-Flag M2 affinity gel at 4 °C. The beads were then washed two times with lysis buffer with 500 mM NaCl and 0.5% NP40 and three times with lysis buffer. The proteins were eluted with one bed volume of 1.25 mg/ml 3X FLAG peptide in 50 mM Tris–HCl, pH 7.5 and 150 mM NaCl for three times and eluates were incubated overnight with 150 μl bed volume of Talon cobalt resin. The resin was then washed five times with buffer containing 50 mM Tris, 150 mM NaCl and 50 mM immidazole and subsequently eluted with 50 mM Tris, 150 mM NaCl and 300 mM immidazole. The eluates were finally incubated with 10% trichloroacetic acid to precipitate the associated proteins overnight at −20 °C and the protein pellets were washed twice with ice-cold acetone and dried.
Mass spectrometry and data analysis
Eluted protein complexes were separated by 1D SDS–PAGE and digested with trypsin using minor variants on published procedures (Shevchenko et al., 2006). Samples were analyzed on an Orbitrap Classic (Thermo Fisher Scientific, Rockford, IL, USA). Survey full scan mass spectrometry (MS) spectra (m/z 400–1400) were acquired with a resolution of R=60 000 at m/z 400, an AGC target of 1e6 ions, and a maximum injection time of 500 ms. The 10 most intense peptide ions in each survey scan with an ion intensity above 1000 counts and a charge state ⩾2 were sequentially isolated to a target value of 1e4 and fragmented in the linear ion trap by collisionally induced dissociation (CID/CAD) using a normalized collision energy of 35%. A dynamic exclusion was applied using a maximum exclusion list of 500 with one repeat count, repeat and exclusion duration of 45 and 30 s, respectively. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK, version 2.0). Mascot was set up to search the IPI.HUMAN.v3.32 database (71 844 sequences) assuming the digestion enzyme trypsin. A fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 5 p.p.m. were used. Iodoacetamide derivative of cysteine was specified as a fixed modification. Pyro-glu from E of glutamic acid, pyro-glu from Q of glutamine, deamidation of asparagine, oxidation of methionine and acetylation of the N-terminus were specified as variable modifications. Scaffold (version Scaffold_2_05_02, Proteome Software Inc, Portland, OR, USA) was used to validate MS/MS-based peptide and protein identifications.
Statistics
Statistical significance was assessed by Student's t-test. A P-value less than 0.05 was considered to be statistically significant.
Other materials and methods
They are described as Supplementary information.
References
Bandura JL, Edgar BA . (2008). Yorkie and scalloped: partners in growth activation. Dev Cell 14: 315–316.
Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W et al. (2008). A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 68: 2592–2598.
Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W . (2009). TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem 284: 14347–14358.
Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S et al. (1997). Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem 272: 17070–17077.
Chen HI, Sudol M . (1995). The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA 92: 7819–7823.
Chen Y, Choong LY, Lin Q, Philp R, Wong CH, Ang BK et al. (2007). Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol Cell Proteomics 6: 2072–2087.
Dhananjayan SC, Ramamoorthy S, Khan OY, Ismail A, Sun J, Slingerland J et al. (2006). WW domain binding protein-2, an E6-associated protein interacting protein, acts as a coactivator of estrogen and progesterone receptors. Mol Endocrinol 20: 2343–2354.
Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA et al. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133.
Einbond A, Sudol M . (1996). Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett 384: 1–8.
Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A . (2008). Scalloped interacts with Yorkie, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol 18: 435–441.
Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ et al. (2008). DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer 47: 490–499.
Harvey K, Tapon N . (2007). The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer 7: 182–191.
Harvey KF, Dinudom A, Komwatana P, Jolliffe CN, Day ML, Parasivam G et al. (1999). All three WW domains of murine Nedd4 are involved in the regulation of epithelial sodium channels by intracellular Na+. J Biol Chem 274: 12525–12530.
Huang J, Wu S, Barrera J, Matthews K, Pan D . (2005). The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122: 421–434.
Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M et al. (2000). TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 19: 6778–6791.
Lam-Himlin DM, Daniels JA, Gayyed MF, Dong J, Maitra A, Pan D et al. (2006). The hippo pathway in human upper gastrointestinal dysplasia and carcinoma: a novel oncogenic pathway. Int J Gastrointest Cancer 37: 103–109.
Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH et al. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426–2436.
Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M et al. (1996). Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382: 646–649.
Mahoney Jr WM, Hong JH, Yaffe MB, Farrance IK . (2005). The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J 388: 217–225.
Nolo R, Morrison CM, Tao C, Zhang X, Halder G . (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol 16: 1895–1904.
Oh H, Irvine KD . (2009). in vivo analysis of Yorkie phosphorylation sites. Oncogene 28: 1916–1927.
Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC et al. (2006). Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA 103: 12405–12410.
Pan D . (2007). Hippo signaling in organ size control. Genes Dev 21: 886–897.
Peng HW, Slattery M, Mann RS . (2009). Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 23: 2307–2319.
Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A et al. (2006). X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9: 121–132.
Saucedo LJ, Edgar BA . (2007). Filling out the hippo pathway. Nat Rev Mol Cell Biol 8: 613–621.
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M . (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860.
Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D et al. (2008). Expression of Yes-associated protein in common solid tumors. Hum Pathol 39: 1582–1589.
Sudol M, Chen HI, Bougeret C, Einbond A, Bork P . (1995). Characterization of a novel protein-binding module--the WW domain. FEBS Lett 369: 67–71.
Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA et al. (2002). Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467–478.
Thompson BJ, Cohen SM . (2006). The hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126: 767–774.
Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML . (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15: 1229–1241.
Wang K, Degerny C, Xu M, Yang XJ . (2009). YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem Cell Biol 87: 77–91.
Wu S, Liu Y, Zheng Y, Dong J, Pan D . (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the hippo growth-regulatory pathway. Dev Cell 14: 388–398.
Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J et al. (2006). Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125: 1253–1267.
Zeng Q, Hong W . (2008). The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13: 188–192.
Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S et al. (2009a). TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem 284: 13355–13362.
Zhang X, Milton CC, Humbert PO, Harvey KF . (2009b). Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res 69: 6033–6041.
Zhao B, Kim J, Ye X, Lai ZC, Guan KL . (2009). Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res 69: 1089–1098.
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761.
Zhao B, Ye X, Yu J, Li L, Li W, Li S et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971.
Acknowledgements
This work is funded by Agency for Science, Technology, and Research (A*STAR), Singapore.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Chan, S., Lim, C., Huang, C. et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene 30, 600–610 (2011). https://doi.org/10.1038/onc.2010.438
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2010.438
Keywords
This article is cited by
-
WBP2 negatively regulates the Hippo pathway by competitively binding to WWC3 with LATS1 to promote non-small cell lung cancer progression
Cell Death & Disease (2021)
-
The emerging roles of WBP2 oncogene in human cancers
Oncogene (2020)
-
Hippo/MST blocks breast cancer by downregulating WBP2 oncogene expression via miRNA processor Dicer
Cell Death & Disease (2020)
-
WW domain-binding protein 2 acts as an oncogene by modulating the activity of the glycolytic enzyme ENO1 in glioma
Cell Death & Disease (2018)
-
Interaction of WBP2 with ERα increases doxorubicin resistance of breast cancer cells by modulating MDR1 transcription
British Journal of Cancer (2018)